
- •COMPUTATIONAL CHEMISTRY
- •CONTENTS
- •PREFACE
- •1.1 WHAT YOU CAN DO WITH COMPUTATIONAL CHEMISTRY
- •1.2 THE TOOLS OF COMPUTATIONAL CHEMISTRY
- •1.3 PUTTING IT ALL TOGETHER
- •1.4 THE PHILOSOPHY OF COMPUTATIONAL CHEMISTRY
- •1.5 SUMMARY OF CHAPTER 1
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •2.1 PERSPECTIVE
- •2.2 STATIONARY POINTS
- •2.3 THE BORN–OPPENHEIMER APPROXIMATION
- •2.4 GEOMETRY OPTIMIZATION
- •2.6 SYMMETRY
- •2.7 SUMMARY OF CHAPTER 2
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •3.1 PERSPECTIVE
- •3.2 THE BASIC PRINCIPLES OF MM
- •3.2.1 Developing a forcefield
- •3.2.2 Parameterizing a forcefield
- •3.2.3 A calculation using our forcefield
- •3.3 EXAMPLES OF THE USE OF MM
- •3.3.2 Geometries and energies of polymers
- •3.3.3 Geometries and energies of transition states
- •3.3.4 MM in organic synthesis
- •3.3.5 Molecular dynamics and Monte Carlo simulations
- •3.4 GEOMETRIES CALCULATED BY MM
- •3.5 FREQUENCIES CALCULATED BY MM
- •3.6 STRENGTHS AND WEAKNESSES OF MM
- •3.6.1 Strengths
- •3.6.2 Weaknesses
- •3.7 SUMMARY OF CHAPTER 3
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •4.1 PERSPECTIVE
- •4.2.1 The origins of quantum theory: blackbody radiation and the photoelectric effect
- •4.2.2 Radioactivity
- •4.2.3 Relativity
- •4.2.4 The nuclear atom
- •4.2.5 The Bohr atom
- •4.2.6 The wave mechanical atom and the Schrödinger equation
- •4.3.1 Introduction
- •4.3.2 Hybridization
- •4.3.3 Matrices and determinants
- •4.3.4 The simple Hückel method – theory
- •4.3.5 The simple Hückel method – applications
- •4.3.6 Strengths and weaknesses of the SHM
- •4.4.1 Theory
- •4.4.2 An illustration of the EHM: the protonated helium molecule
- •4.4.3 The extended Hückel method – applications
- •4.4.4 Strengths and weaknesses of the EHM
- •4.5 SUMMARY OF CHAPTER 4
- •REFERENCES
- •EASIER QUESTIONS
- •5.1 PERSPECTIVE
- •5.2.1 Preliminaries
- •5.2.2 The Hartree SCF method
- •5.2.3 The HF equations
- •5.2.3.1 Slater determinants
- •5.2.3.2 Calculating the atomic or molecular energy
- •5.2.3.3 The variation theorem (variation principle)
- •5.2.3.4 Minimizing the energy; the HF equations
- •5.2.3.5 The meaning of the HF equations
- •5.2.3.6a Deriving the Roothaan–Hall equations
- •5.3 BASIS SETS
- •5.3.1 Introduction
- •5.3.2 Gaussian functions; basis set preliminaries; direct SCF
- •5.3.3 Types of basis sets and their uses
- •5.4 POST-HF CALCULATIONS: ELECTRON CORRELATION
- •5.4.1 Electron correlation
- •5.4.3 The configuration interaction approach to electron correlation
- •5.5.1 Geometries
- •5.5.2 Energies
- •5.5.2.1 Energies: Preliminaries
- •5.5.2.2 Energies: calculating quantities relevant to thermodynamics and to kinetics
- •5.5.2.2a Thermodynamics; “direct” methods, isodesmic reactions
- •5.5.2.2b Thermodynamics; high-accuracy calculations
- •5.5.2.3 Thermodynamics; calculating heats of formation
- •5.5.2.3a Kinetics; calculating reaction rates
- •5.5.2.3b Energies: concluding remarks
- •5.5.3 Frequencies
- •Dipole moments
- •Charges and bond orders
- •Electrostatic potential
- •Atoms-in-molecules
- •5.5.5 Miscellaneous properties – UV and NMR spectra, ionization energies, and electron affinities
- •5.5.6 Visualization
- •5.6 STRENGTHS AND WEAKNESSES OF AB INITIO CALCULATIONS
- •5.7 SUMMARY OF CHAPTER 5
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •6.1 PERSPECTIVE
- •6.2 THE BASIC PRINCIPLES OF SCF SE METHODS
- •6.2.1 Preliminaries
- •6.2.2 The Pariser-Parr-Pople (PPP) method
- •6.2.3 The complete neglect of differential overlap (CNDO) method
- •6.2.4 The intermediate neglect of differential overlap (INDO) method
- •6.2.5 The neglect of diatomic differential overlap (NDDO) method
- •6.2.5.2 Heats of formation from SE electronic energies
- •6.2.5.3 MNDO
- •6.2.5.7 Inclusion of d orbitals: MNDO/d and PM3t; explicit electron correlation: MNDOC
- •6.3 APPLICATIONS OF SE METHODS
- •6.3.1 Geometries
- •6.3.2 Energies
- •6.3.2.1 Energies: preliminaries
- •6.3.2.2 Energies: calculating quantities relevant to thermodynamics and kinetics
- •6.3.3 Frequencies
- •6.3.4 Properties arising from electron distribution: dipole moments, charges, bond orders
- •6.3.5 Miscellaneous properties – UV spectra, ionization energies, and electron affinities
- •6.3.6 Visualization
- •6.3.7 Some general remarks
- •6.4 STRENGTHS AND WEAKNESSES OF SE METHODS
- •6.5 SUMMARY OF CHAPTER 6
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •7.1 PERSPECTIVE
- •7.2 THE BASIC PRINCIPLES OF DENSITY FUNCTIONAL THEORY
- •7.2.1 Preliminaries
- •7.2.2 Forerunners to current DFT methods
- •7.2.3.1 Functionals: The Hohenberg–Kohn theorems
- •7.2.3.2 The Kohn–Sham energy and the KS equations
- •7.2.3.3 Solving the KS equations
- •7.2.3.4a The local density approximation (LDA)
- •7.2.3.4b The local spin density approximation (LSDA)
- •7.2.3.4c Gradient-corrected functionals and hybrid functionals
- •7.3 APPLICATIONS OF DENSITY FUNCTIONAL THEORY
- •7.3.1 Geometries
- •7.3.2 Energies
- •7.3.2.1 Energies: preliminaries
- •7.3.2.2 Energies: calculating quantities relevant to thermodynamics and kinetics
- •7.3.2.2a Thermodynamics
- •7.3.2.2b Kinetics
- •7.3.3 Frequencies
- •7.3.6 Visualization
- •7.4 STRENGTHS AND WEAKNESSES OF DFT
- •7.5 SUMMARY OF CHAPTER 7
- •REFERENCES
- •EASIER QUESTIONS
- •HARDER QUESTIONS
- •8.1 FROM THE LITERATURE
- •8.1.1.1 Oxirene
- •8.1.1.2 Nitrogen pentafluoride
- •8.1.1.3 Pyramidane
- •8.1.1.4 Beyond dinitrogen
- •8.1.2 Mechanisms
- •8.1.2.1 The Diels–Alder reaction
- •8.1.2.2 Abstraction of H from amino acids by the OH radical
- •8.1.3 Concepts
- •8.1.3.1 Resonance vs. inductive effects
- •8.1.3.2 Homoaromaticity
- •8.2 TO THE LITERATURE
- •8.2.1 Books
- •8.2.2 The Worldwide Web
- •8.3 SOFTWARE AND HARDWARE
- •8.3.1 Software
- •8.3.2 Hardware
- •8.3.3 Postscript
- •REFERENCES
- •INDEX

50 Computational Chemistry
Setting  we find that for this point on the curve
we find that for this point on the curve  i.e.
i.e.
If we set  (from Eq. (3.6)) in Eq. (3.5), we find
(from Eq. (3.6)) in Eq. (3.5), we find
i.e.
So  can be calculated from the depth of the energy minimum.
can be calculated from the depth of the energy minimum.
In deciding to use equations of the form (3.2)–(3.5) we have decided on a particular MM forcefield. There are many alternative forcefields. For example, we might have chosen to approximate  by the sum of a quadratic and a cubic term:
by the sum of a quadratic and a cubic term:
This gives a somewhat more accurate representation of the variation of energy with length. Again, we might have represented the nonbonded interaction energy by a more complicated expression than the simple 12-6 potential of Eq. (3.5) (which is by no means the best form for nonbonded repulsions). Such changes would represent changes in the forcefield.
3.2.2 Parameterizing a forcefield
We can now consider putting actual numbers,  etc., into Eqs (3.2)– (3.5), to give expressions that we can actually use. The process of finding these numbers is called parameterizing (or parametrizing) the forcefield. The set of molecules used for parameterization, perhaps 100 for a good forcefield, is called the training set. In the purely illustrative example below we use just ethane, methane and butane.
etc., into Eqs (3.2)– (3.5), to give expressions that we can actually use. The process of finding these numbers is called parameterizing (or parametrizing) the forcefield. The set of molecules used for parameterization, perhaps 100 for a good forcefield, is called the training set. In the purely illustrative example below we use just ethane, methane and butane.
Parameterizing the bond stretching term. A forcefield can be parameterized by reference to experiment (empirical parameterization) or by getting the numbers from high-level ab initio or density functional calculations, or by a combination of both approaches. For the bond stretching term of Eq. (3.2) we need  and Experimentally,
and Experimentally,  could be obtained from IR spectra, as the stretching frequency of a bond depends on the force constant (and the masses of the atoms involved) [8], and
could be obtained from IR spectra, as the stretching frequency of a bond depends on the force constant (and the masses of the atoms involved) [8], and  could be derived from X-ray diffraction, electron diffraction, or microwave spectroscopy [9].
could be derived from X-ray diffraction, electron diffraction, or microwave spectroscopy [9].
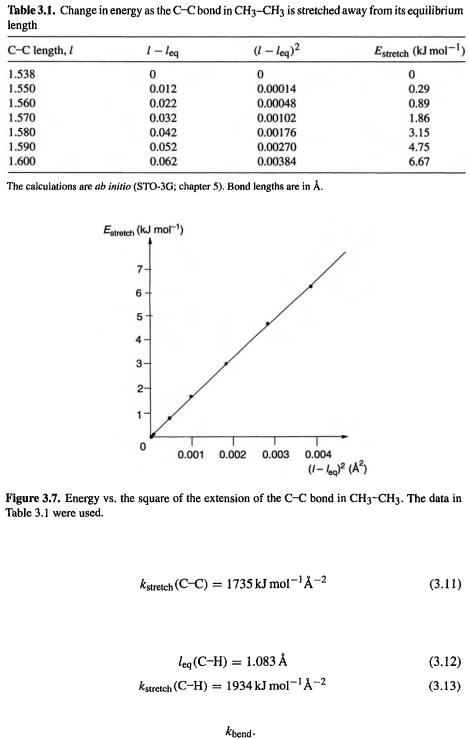
Let us find for the C/C bond of ethane by ab initio (chapter 5) calculations. Normally high-level ab initio calculations would be used to parameterize a forcefield, but for illustrative purposes we can use the low-level but fast STO-3G method [10]. Eq. (3.2) shows that a plot of
for the C/C bond of ethane by ab initio (chapter 5) calculations. Normally high-level ab initio calculations would be used to parameterize a forcefield, but for illustrative purposes we can use the low-level but fast STO-3G method [10]. Eq. (3.2) shows that a plot of  against
against  should be linear with a slope of
should be linear with a slope of  Table 3.1 and Fig. 3.7 show the variation of the energy of ethane with stretching of the C/C bond, as calculated by the ab initio STO-3G method. The equilibrium bond
Table 3.1 and Fig. 3.7 show the variation of the energy of ethane with stretching of the C/C bond, as calculated by the ab initio STO-3G method. The equilibrium bond
length has been taken as the STO-3G length:
Molecular Mechanics 51
The slope of the graph is
Similarly, the CH bond of methane was stretched using ab initio STO-3G calculations; the results are
Parameterizing the angle bending term.  should be linear with a slope of
should be linear with a slope of
From Eq. (3.3), a plot of  against From STO-3G calculations on bending
against From STO-3G calculations on bending

52 Computational Chemistry
the H–C–C angle in ethane we get (cf. Table 3.1 and Fig. 3.7)
Calculations on staggered butane gave for the C–C–C angle
Parameterizing the torsional term. For the ethane case (Fig. 3.4), the equation for energy as a function of dihedral angle can be deduced fairly simply by adjusting the basic equation  to give
to give  For butane (Fig. 3.5), using Eq. (3.4) and experimenting with a curve-fitting program shows that a reasonably accurate torsional potential energy function can be created with five parameters,
For butane (Fig. 3.5), using Eq. (3.4) and experimenting with a curve-fitting program shows that a reasonably accurate torsional potential energy function can be created with five parameters,  and
and
The values of the parameters 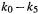 are given in Table 3.2. The calculated curve can be made to match the experimental one as closely as desired by using more terms (Fourier analysis).
are given in Table 3.2. The calculated curve can be made to match the experimental one as closely as desired by using more terms (Fourier analysis).
Parameterizing the nonbonded interactions term. To parameterize Eq. (3.5) we might perform ab initio calculations in which the separation of two atoms or groups in different

Molecular Mechanics 53
molecules (to avoid the complication of concomitant changes in bond lengths and angles) is varied, and fit Eq. (3.5) to the energy vs. distance results. For nonpolar groups this would require quite high-level calculations (chapter 5), as van der Waals or dispersion forces are involved. We shall approximate the nonbonded interactions of methyl groups by the interactions of methane molecules, using experimental values of  and
and  , derived from studies of the viscosity or the compressibility of methane. The two methods give slightly different values [7b], but we can use the values
, derived from studies of the viscosity or the compressibility of methane. The two methods give slightly different values [7b], but we can use the values
and
Summary of the parameterization of the forcefield terms. The four terms of Eq. (3.1) were parameterized to give:
The parameters k of Eq. (3.25) are given in Table 3.2.
Note that this parameterization is only illustrative of the principles involved; any really viable forcefield would actually be much more sophisticated. The kind we have developed here might at the very best give crude estimates of the energies of alkanes. An accurate, practical forcefield would be parameterized as a best fit to many experimental and/or calculational results, and would have different parameters for different kinds of bonds, e.g. C–C for acyclic alkanes, for cyclobutane and for cyclopropane. A forcefield able to handle not only hydrocarbons would obviously need parameters involving elements other than hydrogen and carbon. Practical forcefields also have different parameters for various atom types, like  carbon vs.
carbon vs.  carbon, or amine nitrogen vs. amide nitrogen. In other words, a different value would be used for, say, stretching involving an
carbon, or amine nitrogen vs. amide nitrogen. In other words, a different value would be used for, say, stretching involving an  C–C bond than for an
C–C bond than for an  C–C bond. This is clearly necessary since the force constant of a bond depends on the hybridization of the atoms involved; the IR stretch frequency for the
C–C bond. This is clearly necessary since the force constant of a bond depends on the hybridization of the atoms involved; the IR stretch frequency for the  bond comes at roughly
bond comes at roughly  while that for the
while that for the  bond is about
bond is about 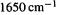 [8]. Since the vibrational frequency of a bond is proportional to the square root of the force constant, the force constants are in the ratio of about
[8]. Since the vibrational frequency of a bond is proportional to the square root of the force constant, the force constants are in the ratio of about  for corresponding atoms, force
for corresponding atoms, force
