
- •brief contents
- •contents
- •preface
- •acknowledgments
- •about this book
- •What’s new in the second edition
- •Who should read this book
- •Roadmap
- •Advice for data miners
- •Code examples
- •Code conventions
- •Author Online
- •About the author
- •about the cover illustration
- •1 Introduction to R
- •1.2 Obtaining and installing R
- •1.3 Working with R
- •1.3.1 Getting started
- •1.3.2 Getting help
- •1.3.3 The workspace
- •1.3.4 Input and output
- •1.4 Packages
- •1.4.1 What are packages?
- •1.4.2 Installing a package
- •1.4.3 Loading a package
- •1.4.4 Learning about a package
- •1.5 Batch processing
- •1.6 Using output as input: reusing results
- •1.7 Working with large datasets
- •1.8 Working through an example
- •1.9 Summary
- •2 Creating a dataset
- •2.1 Understanding datasets
- •2.2 Data structures
- •2.2.1 Vectors
- •2.2.2 Matrices
- •2.2.3 Arrays
- •2.2.4 Data frames
- •2.2.5 Factors
- •2.2.6 Lists
- •2.3 Data input
- •2.3.1 Entering data from the keyboard
- •2.3.2 Importing data from a delimited text file
- •2.3.3 Importing data from Excel
- •2.3.4 Importing data from XML
- •2.3.5 Importing data from the web
- •2.3.6 Importing data from SPSS
- •2.3.7 Importing data from SAS
- •2.3.8 Importing data from Stata
- •2.3.9 Importing data from NetCDF
- •2.3.10 Importing data from HDF5
- •2.3.11 Accessing database management systems (DBMSs)
- •2.3.12 Importing data via Stat/Transfer
- •2.4 Annotating datasets
- •2.4.1 Variable labels
- •2.4.2 Value labels
- •2.5 Useful functions for working with data objects
- •2.6 Summary
- •3 Getting started with graphs
- •3.1 Working with graphs
- •3.2 A simple example
- •3.3 Graphical parameters
- •3.3.1 Symbols and lines
- •3.3.2 Colors
- •3.3.3 Text characteristics
- •3.3.4 Graph and margin dimensions
- •3.4 Adding text, customized axes, and legends
- •3.4.1 Titles
- •3.4.2 Axes
- •3.4.3 Reference lines
- •3.4.4 Legend
- •3.4.5 Text annotations
- •3.4.6 Math annotations
- •3.5 Combining graphs
- •3.5.1 Creating a figure arrangement with fine control
- •3.6 Summary
- •4 Basic data management
- •4.1 A working example
- •4.2 Creating new variables
- •4.3 Recoding variables
- •4.4 Renaming variables
- •4.5 Missing values
- •4.5.1 Recoding values to missing
- •4.5.2 Excluding missing values from analyses
- •4.6 Date values
- •4.6.1 Converting dates to character variables
- •4.6.2 Going further
- •4.7 Type conversions
- •4.8 Sorting data
- •4.9 Merging datasets
- •4.9.1 Adding columns to a data frame
- •4.9.2 Adding rows to a data frame
- •4.10 Subsetting datasets
- •4.10.1 Selecting (keeping) variables
- •4.10.2 Excluding (dropping) variables
- •4.10.3 Selecting observations
- •4.10.4 The subset() function
- •4.10.5 Random samples
- •4.11 Using SQL statements to manipulate data frames
- •4.12 Summary
- •5 Advanced data management
- •5.2 Numerical and character functions
- •5.2.1 Mathematical functions
- •5.2.2 Statistical functions
- •5.2.3 Probability functions
- •5.2.4 Character functions
- •5.2.5 Other useful functions
- •5.2.6 Applying functions to matrices and data frames
- •5.3 A solution for the data-management challenge
- •5.4 Control flow
- •5.4.1 Repetition and looping
- •5.4.2 Conditional execution
- •5.5 User-written functions
- •5.6 Aggregation and reshaping
- •5.6.1 Transpose
- •5.6.2 Aggregating data
- •5.6.3 The reshape2 package
- •5.7 Summary
- •6 Basic graphs
- •6.1 Bar plots
- •6.1.1 Simple bar plots
- •6.1.2 Stacked and grouped bar plots
- •6.1.3 Mean bar plots
- •6.1.4 Tweaking bar plots
- •6.1.5 Spinograms
- •6.2 Pie charts
- •6.3 Histograms
- •6.4 Kernel density plots
- •6.5 Box plots
- •6.5.1 Using parallel box plots to compare groups
- •6.5.2 Violin plots
- •6.6 Dot plots
- •6.7 Summary
- •7 Basic statistics
- •7.1 Descriptive statistics
- •7.1.1 A menagerie of methods
- •7.1.2 Even more methods
- •7.1.3 Descriptive statistics by group
- •7.1.4 Additional methods by group
- •7.1.5 Visualizing results
- •7.2 Frequency and contingency tables
- •7.2.1 Generating frequency tables
- •7.2.2 Tests of independence
- •7.2.3 Measures of association
- •7.2.4 Visualizing results
- •7.3 Correlations
- •7.3.1 Types of correlations
- •7.3.2 Testing correlations for significance
- •7.3.3 Visualizing correlations
- •7.4 T-tests
- •7.4.3 When there are more than two groups
- •7.5 Nonparametric tests of group differences
- •7.5.1 Comparing two groups
- •7.5.2 Comparing more than two groups
- •7.6 Visualizing group differences
- •7.7 Summary
- •8 Regression
- •8.1 The many faces of regression
- •8.1.1 Scenarios for using OLS regression
- •8.1.2 What you need to know
- •8.2 OLS regression
- •8.2.1 Fitting regression models with lm()
- •8.2.2 Simple linear regression
- •8.2.3 Polynomial regression
- •8.2.4 Multiple linear regression
- •8.2.5 Multiple linear regression with interactions
- •8.3 Regression diagnostics
- •8.3.1 A typical approach
- •8.3.2 An enhanced approach
- •8.3.3 Global validation of linear model assumption
- •8.3.4 Multicollinearity
- •8.4 Unusual observations
- •8.4.1 Outliers
- •8.4.3 Influential observations
- •8.5 Corrective measures
- •8.5.1 Deleting observations
- •8.5.2 Transforming variables
- •8.5.3 Adding or deleting variables
- •8.5.4 Trying a different approach
- •8.6 Selecting the “best” regression model
- •8.6.1 Comparing models
- •8.6.2 Variable selection
- •8.7 Taking the analysis further
- •8.7.1 Cross-validation
- •8.7.2 Relative importance
- •8.8 Summary
- •9 Analysis of variance
- •9.1 A crash course on terminology
- •9.2 Fitting ANOVA models
- •9.2.1 The aov() function
- •9.2.2 The order of formula terms
- •9.3.1 Multiple comparisons
- •9.3.2 Assessing test assumptions
- •9.4 One-way ANCOVA
- •9.4.1 Assessing test assumptions
- •9.4.2 Visualizing the results
- •9.6 Repeated measures ANOVA
- •9.7 Multivariate analysis of variance (MANOVA)
- •9.7.1 Assessing test assumptions
- •9.7.2 Robust MANOVA
- •9.8 ANOVA as regression
- •9.9 Summary
- •10 Power analysis
- •10.1 A quick review of hypothesis testing
- •10.2 Implementing power analysis with the pwr package
- •10.2.1 t-tests
- •10.2.2 ANOVA
- •10.2.3 Correlations
- •10.2.4 Linear models
- •10.2.5 Tests of proportions
- •10.2.7 Choosing an appropriate effect size in novel situations
- •10.3 Creating power analysis plots
- •10.4 Other packages
- •10.5 Summary
- •11 Intermediate graphs
- •11.1 Scatter plots
- •11.1.3 3D scatter plots
- •11.1.4 Spinning 3D scatter plots
- •11.1.5 Bubble plots
- •11.2 Line charts
- •11.3 Corrgrams
- •11.4 Mosaic plots
- •11.5 Summary
- •12 Resampling statistics and bootstrapping
- •12.1 Permutation tests
- •12.2 Permutation tests with the coin package
- •12.2.2 Independence in contingency tables
- •12.2.3 Independence between numeric variables
- •12.2.5 Going further
- •12.3 Permutation tests with the lmPerm package
- •12.3.1 Simple and polynomial regression
- •12.3.2 Multiple regression
- •12.4 Additional comments on permutation tests
- •12.5 Bootstrapping
- •12.6 Bootstrapping with the boot package
- •12.6.1 Bootstrapping a single statistic
- •12.6.2 Bootstrapping several statistics
- •12.7 Summary
- •13 Generalized linear models
- •13.1 Generalized linear models and the glm() function
- •13.1.1 The glm() function
- •13.1.2 Supporting functions
- •13.1.3 Model fit and regression diagnostics
- •13.2 Logistic regression
- •13.2.1 Interpreting the model parameters
- •13.2.2 Assessing the impact of predictors on the probability of an outcome
- •13.2.3 Overdispersion
- •13.2.4 Extensions
- •13.3 Poisson regression
- •13.3.1 Interpreting the model parameters
- •13.3.2 Overdispersion
- •13.3.3 Extensions
- •13.4 Summary
- •14 Principal components and factor analysis
- •14.1 Principal components and factor analysis in R
- •14.2 Principal components
- •14.2.1 Selecting the number of components to extract
- •14.2.2 Extracting principal components
- •14.2.3 Rotating principal components
- •14.2.4 Obtaining principal components scores
- •14.3 Exploratory factor analysis
- •14.3.1 Deciding how many common factors to extract
- •14.3.2 Extracting common factors
- •14.3.3 Rotating factors
- •14.3.4 Factor scores
- •14.4 Other latent variable models
- •14.5 Summary
- •15 Time series
- •15.1 Creating a time-series object in R
- •15.2 Smoothing and seasonal decomposition
- •15.2.1 Smoothing with simple moving averages
- •15.2.2 Seasonal decomposition
- •15.3 Exponential forecasting models
- •15.3.1 Simple exponential smoothing
- •15.3.3 The ets() function and automated forecasting
- •15.4 ARIMA forecasting models
- •15.4.1 Prerequisite concepts
- •15.4.2 ARMA and ARIMA models
- •15.4.3 Automated ARIMA forecasting
- •15.5 Going further
- •15.6 Summary
- •16 Cluster analysis
- •16.1 Common steps in cluster analysis
- •16.2 Calculating distances
- •16.3 Hierarchical cluster analysis
- •16.4 Partitioning cluster analysis
- •16.4.2 Partitioning around medoids
- •16.5 Avoiding nonexistent clusters
- •16.6 Summary
- •17 Classification
- •17.1 Preparing the data
- •17.2 Logistic regression
- •17.3 Decision trees
- •17.3.1 Classical decision trees
- •17.3.2 Conditional inference trees
- •17.4 Random forests
- •17.5 Support vector machines
- •17.5.1 Tuning an SVM
- •17.6 Choosing a best predictive solution
- •17.7 Using the rattle package for data mining
- •17.8 Summary
- •18 Advanced methods for missing data
- •18.1 Steps in dealing with missing data
- •18.2 Identifying missing values
- •18.3 Exploring missing-values patterns
- •18.3.1 Tabulating missing values
- •18.3.2 Exploring missing data visually
- •18.3.3 Using correlations to explore missing values
- •18.4 Understanding the sources and impact of missing data
- •18.5 Rational approaches for dealing with incomplete data
- •18.6 Complete-case analysis (listwise deletion)
- •18.7 Multiple imputation
- •18.8 Other approaches to missing data
- •18.8.1 Pairwise deletion
- •18.8.2 Simple (nonstochastic) imputation
- •18.9 Summary
- •19 Advanced graphics with ggplot2
- •19.1 The four graphics systems in R
- •19.2 An introduction to the ggplot2 package
- •19.3 Specifying the plot type with geoms
- •19.4 Grouping
- •19.5 Faceting
- •19.6 Adding smoothed lines
- •19.7 Modifying the appearance of ggplot2 graphs
- •19.7.1 Axes
- •19.7.2 Legends
- •19.7.3 Scales
- •19.7.4 Themes
- •19.7.5 Multiple graphs per page
- •19.8 Saving graphs
- •19.9 Summary
- •20 Advanced programming
- •20.1 A review of the language
- •20.1.1 Data types
- •20.1.2 Control structures
- •20.1.3 Creating functions
- •20.2 Working with environments
- •20.3 Object-oriented programming
- •20.3.1 Generic functions
- •20.3.2 Limitations of the S3 model
- •20.4 Writing efficient code
- •20.5 Debugging
- •20.5.1 Common sources of errors
- •20.5.2 Debugging tools
- •20.5.3 Session options that support debugging
- •20.6 Going further
- •20.7 Summary
- •21 Creating a package
- •21.1 Nonparametric analysis and the npar package
- •21.1.1 Comparing groups with the npar package
- •21.2 Developing the package
- •21.2.1 Computing the statistics
- •21.2.2 Printing the results
- •21.2.3 Summarizing the results
- •21.2.4 Plotting the results
- •21.2.5 Adding sample data to the package
- •21.3 Creating the package documentation
- •21.4 Building the package
- •21.5 Going further
- •21.6 Summary
- •22 Creating dynamic reports
- •22.1 A template approach to reports
- •22.2 Creating dynamic reports with R and Markdown
- •22.3 Creating dynamic reports with R and LaTeX
- •22.4 Creating dynamic reports with R and Open Document
- •22.5 Creating dynamic reports with R and Microsoft Word
- •22.6 Summary
- •afterword Into the rabbit hole
- •appendix A Graphical user interfaces
- •appendix B Customizing the startup environment
- •appendix C Exporting data from R
- •Delimited text file
- •Excel spreadsheet
- •Statistical applications
- •appendix D Matrix algebra in R
- •appendix E Packages used in this book
- •appendix F Working with large datasets
- •F.1 Efficient programming
- •F.2 Storing data outside of RAM
- •F.3 Analytic packages for out-of-memory data
- •F.4 Comprehensive solutions for working with enormous datasets
- •appendix G Updating an R installation
- •G.1 Automated installation (Windows only)
- •G.2 Manual installation (Windows and Mac OS X)
- •G.3 Updating an R installation (Linux)
- •references
- •index
- •Symbols
- •Numerics
- •23.1 The lattice package
- •23.2 Conditioning variables
- •23.3 Panel functions
- •23.4 Grouping variables
- •23.5 Graphic parameters
- •23.6 Customizing plot strips
- •23.7 Page arrangement
- •23.8 Going further
428 |
CHAPTER 18 Advanced methods for missing data |
deletion reduced the sample size by 32%. Next, we’ll consider an approach that employs the entire dataset (including cases with missing data).
18.7 Multiple imputation
Multiple imputation (MI) provides an approach to missing values that’s based on repeated simulations. MI is frequently the method of choice for complex missing-val- ues problems. In MI, a set of complete datasets (typically 3 to 10) is generated from an existing dataset that’s missing values. Monte Carlo methods are used to fill in the missing data in each of the simulated datasets. Standard statistical methods are applied to each of the simulated datasets, and the outcomes are combined to provide estimated results and confidence intervals that take into account the uncertainty introduced by the missing values. Good implementations are available in R through the Amelia, mice, and mi packages.
In this section, we’ll focus |
with() |
|
||
on the approach provided by |
mice() |
pool() |
||
the mice (multivariate impu- |
||||
|
|
|||
tation by chained equations) |
|
|
||
package. To understand how |
|
|
||
the mice package |
operates, |
Data frame |
Final result |
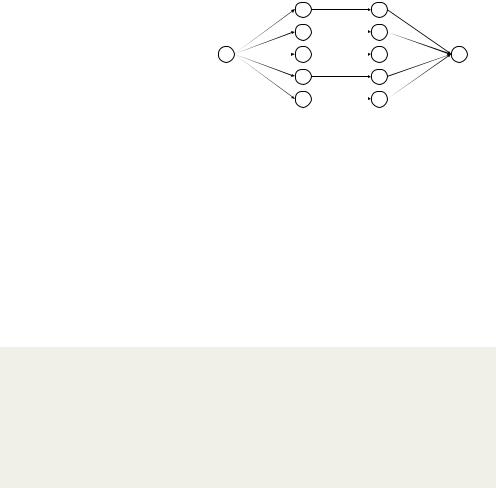
|
consider the diagram in figure |
|
|||
|
|
|||
18.5. |
|
|
|
|
The function mice() starts |
Imputed datasets |
Analysis results |
||
|
|
|||
with a data frame that’s miss- |
Figure 18.5 Steps in applying multiple imputation to missing |
|||
ing data and returns an |
data via the mice approach |
|
||
object containing |
several |
|
|
|
complete datasets (the default is five). Each complete dataset is created by imputing values for the missing data in the original data frame. There’s a random component to the imputations, so each complete dataset is slightly different. The with() function is then used to apply a statistical model (for example, a linear or generalized linear model) to each complete dataset in turn. Finally, the pool() function combines the results of these separate analyses into a single set of results. The standard errors and p-values in this final model correctly reflect the uncertainty produced by both the missing values and the multiple imputations.
How does the mice() function impute missing values?
Missing values are imputed by Gibbs sampling. By default, each variable with missing values is predicted from all other variables in the dataset. These prediction equations are used to impute plausible values for the missing data. The process iterates until convergence over the missing values is achieved. For each variable, you can choose the form of the prediction model (called an elementary imputation method) and the variables entered into it.

Multiple imputation |
429 |
By default, predictive mean matching is used to replace missing data on continuous variables, whereas logistic or polytomous logistic regression is used for target variables that are dichotomous (factors with two levels) or polytomous (factors with more than two levels), respectively. Other elementary imputation methods include Bayesian linear regression, discriminant function analysis, two-level normal imputation, and random sampling from observed values. You can supply your own methods as well.
An analysis based on the mice package typically conforms to the following structure
library(mice)
imp <- mice(data, m)
fit <- with(imp, analysis) pooled <- pool(fit) summary(pooled)
where
■data is a matrix or data frame containing missing values.
■imp is a list object containing the m imputed datasets, along with information on how the imputations were accomplished. By default, m = 5.
■analysis is a formula object specifying the statistical analysis to be applied to each of the m imputed datasets. Examples include lm() for linear regression models, glm() for generalized linear models, gam() for generalized additive models, and nbrm() for negative binomial models. Formulas within the parentheses give the response variables on the left of the ~ and the predictor variables (separated by + signs) on the right.
■fit is a list object containing the results of the m separate statistical analyses.
■pooled is a list object containing the averaged results of these m statistical analyses.
Let’s apply multiple imputation to the sleep dataset. You’ll repeat the analysis from section 18.6, but this time use all 62 mammals. Set the seed value for the random number generator to 1,234 so that your results will match the following:
>library(mice)
>data(sleep, package="VIM")
>imp <- mice(sleep, seed=1234)
[...output deleted to save space...]
>fit <- with(imp, lm(Dream ~ Span + Gest))
>pooled <- pool(fit)
>summary(pooled)
|
est |
|
se |
t |
df Pr(>|t|) |
lo 95 |
|
(Intercept) |
2.58858 |
0.27552 |
9.395 |
52.1 |
8.34e-13 |
2.03576 |
|
Span |
-0.00276 |
0.01295 |
-0.213 52.9 |
8.32e-01 -0.02874 |
|||
Gest |
-0.00421 |
0.00157 |
-2.671 55.6 |
9.91e-03 -0.00736 |
|||
|
hi 95 |
nmis |
|
fmi |
|
|
|
(Intercept) |
3.14141 |
NA |
0.0870 |
|
|
|
|
Span |
0.02322 |
4 |
0.0806 |
|
|
|
|
Gest |
-0.00105 |
4 |
0.0537 |
|
|
|
|
430 |
CHAPTER 18 Advanced methods for missing data |
Here, you see that the regression coefficient for Span isn’t significant (p 0.08), and the coefficient for Gest is significant at the p < 0.01 level. If you compare these results with those produced by a complete case analysis (section 18.6), you see that you’d come to the same conclusions in this instance. Length of gestation has a (statistically) significant, negative relationship with amount of dream sleep, controlling for life span. Although the complete-case analysis was based on the 42 mammals with complete data, the current analysis is based on information gathered from the full set of 62 mammals. By the way, the fmi column reports the fraction of missing information (that is, the proportion of variability that is attributable to the uncertainty introduced by the missing data).
You can access more information about the imputation by examining the objects created in the analysis. For example, let’s view a summary of the imp object:
> imp |
|
|
|
|
|
|
|
|
|
|
|
|
Multiply imputed |
|
data set |
|
|
|
|
|
|
|
|
||
Call: |
|
|
|
|
|
|
|
|
|
|
|
|
mice(data = sleep, seed |
= |
1234) |
|
|
|
|
|
|
||||
Number of multiple imputations: 5 |
|
|
|
|
|
|
||||||
Missing cells per column: |
|
|
|
|
|
|
|
|
||||
BodyWgt BrainWgt |
NonD |
|
Dream |
Sleep |
Span |
|
Gest |
Pred |
||||
0 |
|
0 |
|
14 |
|
12 |
|
4 |
4 |
|
4 |
0 |
Exp |
Danger |
|
|
|
|
|
|
|
|
|
|
|
0 |
|
0 |
|
|
|
|
|
|
|
|
|
|
Imputation methods: |
|
|
|
|
|
|
|
|
|
|||
BodyWgt BrainWgt |
NonD |
|
Dream |
Sleep |
Span |
|
Gest |
Pred |
||||
"" |
"" |
"pmm" |
|
"pmm" |
"pmm" |
"pmm" |
|
"pmm" |
"" |
|||
Exp |
Danger |
|
|
|
|
|
|
|
|
|
|
|
"" |
"" |
|
|
|
|
|
|
|
|
|
|
|
VisitSequence: |
|
|
|
|
|
|
|
|
|
|
|
|
NonD Dream Sleep |
Span |
Gest |
|
|
|
|
|
|
|
|||
3 |
4 |
5 |
6 |
|
7 |
|
|
|
|
|
|
|
PredictorMatrix: |
|
|
|
|
|
|
|
|
|
|
|
|
|
BodyWgt |
|
BrainWgt |
NonD Dream Sleep |
Span |
Gest Pred Exp Danger |
||||||
BodyWgt |
0 |
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
BrainWgt |
0 |
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
NonD |
1 |
|
|
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
Dream |
1 |
|
|
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
Sleep |
1 |
|
|
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
Span |
1 |
|
|
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
Gest |
1 |
|
|
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
Pred |
0 |
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Exp |
0 |
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Danger |
0 |
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Random generator |
|
seed value: |
1234 |
|
|
|
|
|
|
|||
From the resulting output, you can see that five synthetic datasets were created and that the predictive mean matching (pmm) method was used for each variable with missing data. No imputation ("") was needed for BodyWgt, BrainWgt, Pred, Exp, or Danger, because they had no missing values. The visit sequence tells you that variables
Multiple imputation |
431 |
were imputed from right to left, starting with NonD and ending with Gest. Finally, the predictor matrix indicates that each variable with missing data was imputed using all the other variables in the dataset. (In this matrix, the rows represent the variables being imputed, the columns represent the variables used for the imputation, and 1s/0s indicate used/not used).
You can view the imputations by looking at subcomponents of the imp object. For example,
> |
imp$imp$Dream |
|
|
||
|
1 |
2 |
3 |
4 |
5 |
1 |
0.5 |
0.5 |
0.5 |
0.5 |
0.0 |
32.3 2.4 1.9 1.5 2.4
41.2 1.3 5.6 2.3 1.3
140.6 1.0 0.0 0.3 0.5
241.2 1.0 5.6 1.0 6.6
261.9 6.6 0.9 2.2 2.0
301.0 1.2 2.6 2.3 1.4
315.6 0.5 1.2 0.5 1.4
470.7 0.6 1.4 1.8 3.6
530.7 0.5 0.7 0.5 0.5
550.5 2.4 0.7 2.6 2.6
621.9 1.4 3.6 5.6 6.6
displays the 5 imputed values for each of the 12 mammals with missing data on the Dream variable. A review of these matrices helps you determine whether the imputed values are reasonable. A negative value for length of sleep might give you pause (or nightmares).
You can view each of the m imputed datasets via the complete() function. The format is
complete(imp, action=#)
where # specifies one of the m synthetically complete datasets. For example,
>dataset3 <- complete(imp, action=3)
>dataset3
|
BodyWgt BrainWgt |
NonD Dream Sleep Span Gest Pred Exp Danger |
||||||||
1 |
6654.00 |
5712.0 |
2.1 |
0.5 |
3.3 |
38.6 |
645 |
3 |
5 |
3 |
2 |
1.00 |
6.6 |
6.3 |
2.0 |
8.3 |
4.5 |
42 |
3 |
1 |
3 |
3 |
3.38 |
44.5 |
10.6 |
1.9 |
12.5 |
14.0 |
60 |
1 |
1 |
1 |
4 |
0.92 |
5.7 |
11.0 |
5.6 |
16.5 |
4.7 |
25 |
5 |
2 |
3 |
5 |
2547.00 |
4603.0 |
2.1 |
1.8 |
3.9 |
69.0 |
624 |
3 |
5 |
4 |
6 |
10.55 |
179.5 |
9.1 |
0.7 |
9.8 |
27.0 |
180 |
4 |
4 |
4 |
[...output deleted |
to save space...] |
|
|
|
|
|
||||
displays the third (out of five) complete dataset created by the multiple imputation process.
Due to space limitations, we’ve only briefly considered the MI implementation provided in the mice package. The mi and Amelia packages also contain valuable
