
John Wiley & Sons - 2004 - Analysis of Genes and Genomes
.pdf
4.9 REAL-TIME PCR |
179 |
|
|
PCR, the single DNA strand is made double stranded through the binding of another, complementary, primer and the action of Taq DNA polymerase.
Like other methods of mRNA analysis, such as northern blots and nuclease protection assays, RT –PCR can be used to quantify the amount of mRNA that was contained in the original sample. This type of analysis is particularly important for monitoring changes in gene expression. However, because PCR amplification is exponential, small sample-to-sample concentration and loading differences are amplified as well. Even large differences in target concentration (100-fold or more) may produce the same intensity of band after 25 or 30 PCR cycles. Therefore, RT –PCR requires careful optimization when used for quantitative mRNA analysis. Quantitation usually takes one of two forms – relative or absolute.
•Relative quantitation compares transcript abundance across multiple samples, using a co-amplified internal control for sample normalization. Results are expressed as ratios of the gene specific signal to the internal control signal. This yields a corrected relative value for the gene specific product in each sample. These values may be compared between samples for an estimate of the relative expression of target RNA in the samples.
•Absolute quantitation, using competitive RT –PCR, measures the absolute amount (e.g. 5.3 × 105 copies) of a specific mRNA sequence within a sample. Dilutions of a synthetic RNA (containing identical primer binding sites, but slightly shorter than the target RNA) are added to the sample and are co-amplified with the target. The PCR product from the endogenous transcript is then compared with the concentration curve created by the synthetic competitor RNA.
4.9Real-time PCR
Quantitative real-time RT –PCR combines the best attributes of both relative and competitive RT –PCR in that it is accurate, precise, high throughput and relatively easy to perform. Real-time PCR automates the otherwise laborious process of relative RT –PCR by quantitating reaction products for each sample in every cycle. Real-time PCR systems rely upon the detection and quantitation of a fluorescent reporter, whose signal increases in direct proportion to the amount of PCR product in a reaction. In the simplest form, the reporter is the double-strand DNA-specific dye SYBR Green (Wittwer et al., 1997). SYBR Green binds double-stranded DNA, probably in the minor groove, and, upon excitation, emits light. Thus, if the dye is included in a PCR reaction, as a
180 |
POLYMERASE CHAIN REACTION 4 |
|
|
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3′ |
||||
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
Primer binding |
||||||||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
3′ |
|
|
|
|
|
5′ |
3′ |
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
Q |
|
R |
Primer 1 |
|||||||||||
|
|
|
|
|
|
Probe |
|
|
|
|
|
|
|
|
||||||
Primer 2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
5′ |
|
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
Displacement |
||||||||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3′ |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
3′ |
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
Q |
R |
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||||
|
|
|
|
|
Cleavage |
|
|
|||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3′ |
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
3′ |
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
Q |
|
|
|
|
|
R |
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||||
|
|
|
|
|
Completed |
|
|
|
||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
synthesis |
|
|
|
|
||||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3′ |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
3′ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5′ |
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
QR
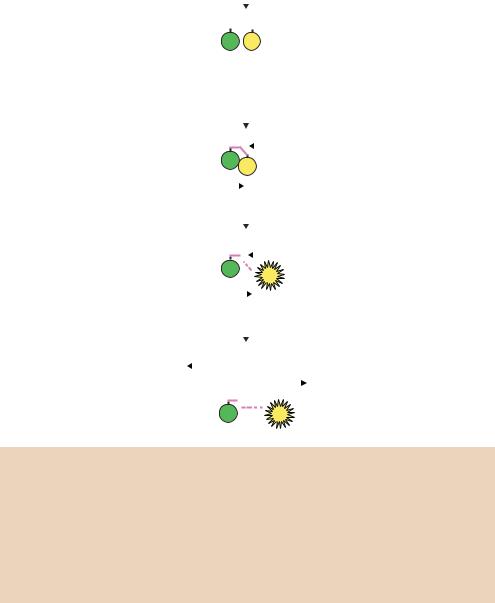
Figure 4.13. TaqMan real-time PCR quantification. Three primers are used during the PCR process – two of these (primers 1 and 2) dictate the beginning of DNA replication on each DNA strand, and the third (the probe) binds to one strand in between. The probe contains two modified bases – a fluorescent reporter (R) at its 5 -end and a fluorescence quencher (Q) at its 3 -end. As DNA replication proceeds, the extended product from primer 1 displaces the 5 -end of the probe and the exonuclease activity of the polymerase cleaves the fluorescent reporter from the probe. The separation of the reporter from the quencher allows it to fluoresce. The amount of fluorescence is proportional to the amount of PCR product being made and is measured during each PCR cycle
PCR product accumulates the fluorescence increases. The advantages of SYBR Green are that it is inexpensive, easy to use, and sensitive. The disadvantage is that SYBR Green will bind to any double-stranded DNA in the reaction, including primer dimers and other non-specific reaction products, which can
4.10 APPLICATIONS OF PCR |
181 |
|
|
result in an over-estimation of the target concentration. For single PCR product reactions with well designed primers, SYBR Green can work extremely well, with spurious non-specific background only showing up in very late cycles.
The alternative method for quantifying PCR products is TaqMan , which relies on fluorescence resonance energy transfer (FRET) of hybridization probes for quantitation (Figure 4.13). TaqMan probes are oligonucleotides that contain a fluorescent reporter dye, typically attached to the 5 base, and a quenching dye, typically attached to the 3 base. The probe is designed to hybridize to an internal region of a PCR product. When irradiated, the excited reporter dye transfers energy to the nearby quenching dye molecule rather than fluorescing, resulting in a non-fluorescent substrate. During PCR, when the polymerase replicates a template on which a probe is bound, the 5 -3 exonuclease activity of the polymerase cleaves the probe (Holland et al., 1991). This separates the fluorescent and quenching dyes and FRET no longer occurs. Fluorescence increases in each PCR cycle, proportional to the rate of probe cleavage, and is measured in a modified thermocycler. Real-time PCR is a powerful quantitative tool, but the cost of reagents and equipment is much higher than that of standard PCR reactions.
4.10Applications of PCR
The polymerase chain reaction has revolutionized molecular biology by allowing the amplification and characterization of minute amounts of nucleic acids. As well as being of use to basic scientists, this technique is of immense importance in medicine for the identification of mutations within small amounts of human DNA, and to pathologists, who routinely need to detect and characterize small amounts of infectious micro-organisms. A detailed description of the many and varied uses to which PCR has been applied is beyond the scope of this text, but a few of the major applications of PCR are listed below:
•molecular cloning
•DNA sequencing
•archaeology
•forensics
•amplification of unknown sequences
•clinical pathology
•genetic diagnosis
•characterizing unknown mutations
182 |
POLYMERASE CHAIN REACTION 4 |
|
|
•fingerprinting/population analysis
•genome analysis
•quantitative PCR of RNA or DNA.
We will touch on some of these topics in later chapters but, again, interested readers are directed toward more dedicated literature (McPherson and Møller, 2000; Innis, Gelfand and Sninsky, 1999).

5.1 GENOMIC LIBRARIES |
185 |
|
|
from PCR products) are becoming increasingly important in genetic engineering experiments.
A genomic DNA library should contain representative copies of all the genetic material of an individual organism. Libraries such as this are organism specific. That is, a library constructed from any tissue from within a single organism should contain the same DNA fragments as those derived from any other tissue. However, libraries generated from different organisms, e.g. those derived from mouse and rat, will be different. Genomic DNA libraries should contain all of the genetic material, whether that material is expressed in a particular tissue type or developmental stage or not. Genomic libraries will therefore contain all DNA sequences: expressed genes, non-expressed genes, exons and introns, promoter and terminator regions and intervening DNA sequences.
cDNA libraries are constructed by the conversion of mRNA from a particular tissue sample into DNA fragments that can be cloned into an appropriate vector. cDNA libraries thus contain only the coding sequence of genes expressed in a tissue sample together with small regions of the 5 and 3 untranslated portions of the gene. Consequently, cDNA libraries isolated from different tissues of the same organism may be radically different in their composition. The genes expressed in one tissue type or developmental stage may well be different from those expressed in another tissue type or developmental stage. Additionally, the composition of a cDNA library reflects the relative abundance of mRNA in the original tissue sample. Highly expressed genes will be represented in the library multiple times, whereas genes expressed at a low level will be represented in the library less frequently.
5.1Genomic Libraries
The smallest unit of DNA within a genome is the chromosome. Even in the simplest organisms, however, chromosomes contain an enormous quantity of DNA. For example, the E. coli chromosome contains some 4.6 Mbp (4 600 000 bp) of DNA (Table 5.1). This amount of DNA is far too large to be cloned into any of the vectors currently available (Chapter 3). Therefore it is necessary, and indeed desirable, to fragment the DNA before it is cloned into an appropriate vector. A ‘divide and conquer’ strategy comes into play here, whereby relatively small fragments of the genome can be assigned a specific function whereas the whole genome is somewhat impenetrable. The method of fragmentation plays an important role in the quality of the final library. Ideally, the genomic DNA should be broken up into random and overlapping fragments prior to cloning. Such cleavage would ensure that the library contains representative copies of all DNA fragments present within the genome, and that fragment

186 CLONING A GENE 5
bias is not encountered by the cleavage of DNA at specific sites only. There are two basic mechanisms for cleaving DNA that are used in the construction of genomic libraries.
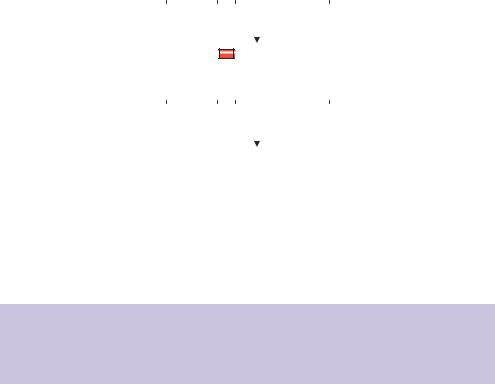
(a)Mechanical shearing. Purified genomic DNA is either passed several times through an narrow-gauge syringe needle or subjected to sonication to break up the DNA into suitable size fragments that can be cloned. Typically, an average DNA fragment size of about 20 kbp is desirable for cloning into λ based vectors. Mechanical methods such as these have the advantage that DNA fragmentation is random, but suffer from the fact that large quantities of DNA are required, and that the average DNA fragmentation size may be quite variable.
(b)Restriction enzyme digestion. Restriction enzymes, such as EcoRI, often recognize 6 bp DNA sequences and cleave the DNA within the recognition sequence. On average, a 6 bp DNA sequence will occur approximately every 4000 bp within DNA. Complete digestion of genomic DNA with EcoRI will generate DNA fragments that are generally too small to be useful in genomic library construction. Other restriction enzymes, e.g. NotI, recognize and cleave 8 bp recognition sequences. Such sequences will occur much less commonly within DNA (approximately once every 65 kbp). However, restriction enzyme cleavage to produce DNA fragments suffers as a consequence of the recognition sites themselves. If, by chance, a gene that we would like to clone contains multiple recognition sites for a particular restriction enzyme, then the fragments generated after enzyme digestion may be too small to clone, and consequently the gene may not be represented within a library. To overcome this problem genomic DNA libraries are usually constructed by digesting the genomic DNA with restriction enzymes in such a way that the digestion does not go to completion (Figure 5.1). Partial restriction digests will ensure that not all DNA recognition sequences are cut and, consequently, that the library produced should contain copies of genes that may possess multiple restriction enzyme recognition sequences. In practice, restriction digestion is normally performed using a restriction enzyme, or often two, that recognize and cleave very commonly occurring sequences. For example, as shown in Figure 5.2, high-molecular-weight genomic DNA is partially cleaved with a mixture of the restriction enzymes HaeIII and AluI. Each of these restriction enzymes recognizes a 4 bp DNA sequence. Their recognition sequences should therefore occur, on average, approximately every 256 bp within genomic DNA. The partial digestion, however, limits the number of restriction enzymes sites that are actually cut and leads to
|
|
|
|
5.1 |
|
GENOMIC LIBRARIES |
187 |
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
(a) |
RE |
RE RE |
RE |
|
||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Complete digest |
|
|
|||
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
(b) |
RE |
RE RE |
RE |
|
||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Partial digest |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Figure 5.1. The complete and partial digestion of a DNA fragment using a restriction enzyme. (a) Complete digestion ensures that all restriction enzyme recognition sites (RE) are cut. (b) Partial digestion results in the cleavage of a random subset of the recognition sites. Partial digestion will generate a variety of products as indicated
the formation of genomic DNA fragments of a suitable size for cloning. DNA fragments produced in this manner have blunt ends since both HaeIII and AluI cut DNA in a blunt-ended fashion:
HaeIII: 5'-GG |
|
CC-3' |
AluI: 5'-AG |
|
CT-3' |
3'-CC |
|
GG-5' |
3'-TC |
|
GA-5' |
|
|
The blunt ended DNA fragments can prove problematical when attempts are made to clone them. As we have already seen (Chapter 2), the ligation of sticky-ended DNA is considerably more efficient than that in which the DNA ends are blunt. Consequently, it is desirable to generate genomic fragments that contain sticky ends in the cloning process. This can be achieved in one of two ways.
•Linkers or adaptors. As shown in Figure 5.2, the blunt ended DNA fragments can be ligated to a series of oligonucleotides that either contain the recognition sequence for a restriction enzyme (linkers) or possess one blunt end for ligation to the genomic DNA and an overhanging sticky end for cloning into particular restriction sites (adaptors). In the case shown here,
