
John Wiley & Sons - 2004 - Analysis of Genes and Genomes
.pdf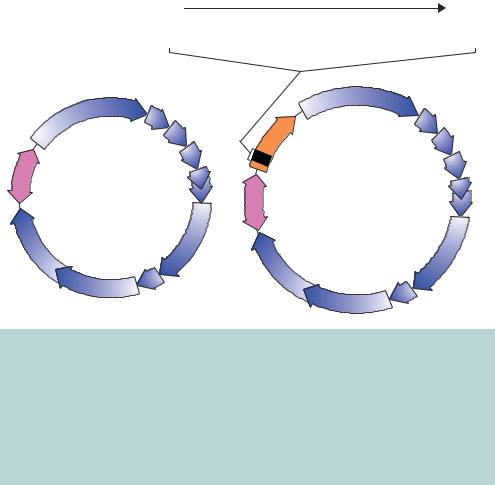
138 |
VECTORS 3 |
|
|
lacZ
(M) T M I T N S |
S S V P |
G D P |
L E S T C |
R H A |
S L A |
||||
EcoRI |
|
KpnI |
BamHI |
|
SalI |
|
|
SphI |
|
5'-atgaccatgattacGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGcact-3' 3'-tactggtactaatgCTTAAGCTCGAGCCATGGGCCCCTAGGAGATCTCAGCTGGACGTCCGTACGTTCGAACCgtga-5'
SacI |
|
SmaI |
|
XbaI |
|
PstI |
HindIII |
Multiple cloning site
2
10
f1 ori |
M13 |
wild-type 6407 bp
2
10 |
5 |
|
lacZ′ |
|
5 |
7 |
|
|
|
|
|
7 |
|
9 |
|
|
|
|
|
9 |
|
8 |
|
M13mp18 |
|
f1 ori |
|
||
|
7250 bp |
8 |
|
|
|
4 |
|
|
3 |
|
|
3 |
|
|
|
|
|
|
|
6 |
4 |
|
1 |
|
|
|
|
6 |
|
|
|
|
|
|
|
|
1 |
Figure 3.15. The genomes of wild-type M13 and an engineered derivative, M13mp18. The wild-type M13 genome encodes 10 open reading frames that are all transcribed in the clockwise direction. Replication of the genome initiates bi-directionally from a specific sequence between genes 2 and 4. M13mp18 additionally bears the lacZ gene for blue–white screening of recombinants. Embedded within this gene is a multiple cloning site providing a number of unique restriction enzyme recognition site to aid the cloning of foreign DNA fragments into the vector. The maps shown here represent the doublestranded from, or replicative form (RF), of the vector that exists within the E. coli host. Viral particles contain only a single strand of the DNA
cell lysis occurring. Up to 1000 phage particles can be released into the medium per cell per generation. M13 phage infection does not result in bacterial cell death and, consequently, M13 infections appear as turbid plaques. The E. coli cells around the site of infection have not been killed, but they grow more slowly due to the burden placed upon them by producing phage particles. The M13 origin of replication (called the f1 ori) contains two overlapping, but distinct, DNA sequences that act to control the synthesis of DNA. These sites – the f1 initiator and the f1 terminator – signal the beginning and end of DNA replication. The initiator is recognized by the protein product of gene 2, which nicks the + strand in the RF DNA. The nick indicates the position at which unidirectional rolling-circle DNA replication will commence. The newly formed + strand is cleaved at the terminator sequence, again by the protein

3.5 M13 VECTORS |
139 |
|
|
|
Single-stranded |
Bacterium |
M13 phage DNA |
|
|
|
F pilus |
|
Protein |
|
coat |
|
M13 |
|
infection |
Viral DNA strand enters bacterium
Conversion to double-stranded RF form
Transcription of viral proteins
Rolling circle replication to form new + strands
Phage assembly
Phage release

140 |
VECTORS 3 |
|
|
product of gene 2. Following cleavage, the two ends of the + strand are ligated to form the single-stranded genome.
The switch between the double-stranded RF form and the single-stranded + form of the M13 viral genome made it an ideal candidate for exploitation as a vector. As we will see in later chapters, the single-stranded DNA produced in the phage particle have led to great advances in mutagenesis in vitro (Chapter 7) and DNA sequencing (Chapter 8). Unlike λ, M13 does not have a non-essential region that can be deleted prior to the insertion of foreign DNA. However, there is an intergenic region between the origin of replication and gene 2 (Figure 3.15) into which foreign DNA fragments may be inserted. M13 vectors were developed in the late 1970s when the lacZ gene (encoding the α-peptide of β-galactosidase) was inserted into the M13 genome (Messing et al., 1977). Subsequently, the same polylinker and α-peptide fragments as the pUC plasmid series were engineered into M13 and naturally occurring restriction enzyme recognition sites were eliminated (Yanisch-Perron, Vieira and Messing, 1985; Norrander, Kempe and Messing, 1983). The RF form of M13 vectors can be isolated by standard plasmid DNA preparation procedures and foreign DNA can be inserted into them as if they were conventional plasmids.
The specific use of M13 vectors is as an aid to the formation of singlestranded DNA. Once a foreign DNA fragment has been cloned into M13, large amounts of the single-stranded form can be easily isolated from the mature phage that are extruded from infected E. coli cells. The main difficulty with vectors of this type is that they tend to be unstable when DNA fragments larger than a few kilobases are inserted into them (Zinder and Boeke, 1982).
3.6Phagemids
Phagemids are plasmids that contain the f1 phage origin of replication for the production of single-stranded DNA. Phagemids are generally small plasmids so that they have the ability to accept larger DNA inserts than M13-based vectors. Phagemids were originally developed in the early 1980s, when it was found
Figure 3.16. The M13 life cycle. The single-stranded M13 genome is encased by coat proteins. Bacterial infection occurs when the phage particle attaches to the E. coli pilus and the single DNA strand is injected into the host. The DNA is immediately converted to a double-stranded form and is replicated and transcribed to produce viral proteins. The build-up of viral protein 2 eventually forces asymmetric DNA replication to produce single DNA strands. These are packaged into new viral particles, which are secreted from the bacteria without cell lysis occurring
3.6 PHAGEMIDS |
141 |
|
|
that the insertion of the f1 origin of replication could be cloned into pBR322 to drive the production of single-stranded DNA (Dotto and Horiuchi, 1981; Dotto, Enea and Zinder, 1981). The f1 replication origin was not sufficient to direct single-stranded DNA production, but if a bacterium carrying a phagemid was superinfected with a functional wild-type M13 or f1 helper phage, then the production of single-stranded phagemid DNA would occur. The phagemid single-stranded DNA would be packaged into viral particles and secreted into the surrounding medium in the same way that M13 phage particles are produced. Additionally, it was found that cloning the f1 origin in the reverse orientation would lead to the production of the opposite strand of DNA (Dente, Cesaveni and Cortese, 1983). Thus, single-stranded DNA representing either strand of a cloned fragment could be produced after cloning into a suitable phagemid vector. Phagemids have the advantage that, in the absence of a helper phage, double-stranded DNA can be isolated as a normal plasmid. Moreover, the lack of additional phage genes the vectors need to carry means that their small size has an increased capacity for carrying larger foreign DNA fragments.
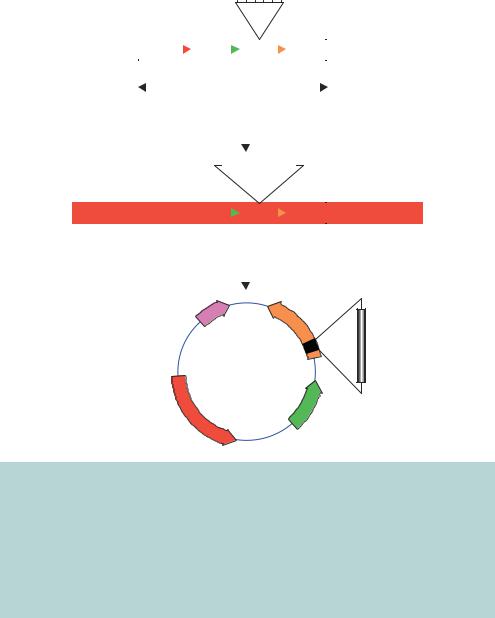
Other phagemids have been developed that take advantage of various aspects of plasmids, λ phage and M13 phage. We have already seen that the f1 replication origin is composed of an initiator and a terminator. In the wild-type M13 phage genome these sequences overlap with each other, such that replication initiates and then terminates after the full circular genome has been replicated. The initiator and terminator elements may be separated from each other to provide starting and ending points for DNA replication on a linear DNA molecule. A λ insertional vector, λZAP, was constructed such that the left-hand and right-hand λ arms were connected via the DNA sequence of a phagemid beginning with the f1 initiator and ending with the f1 terminator (Short et al., 1988). This vector (shown in Figure 3.17) has the ability to function as a λ phage for, for example, the construction of a cDNA library. However, the foreign DNA can be excised from the λ phage in the form of a plasmid after superinfection with a wild-type M13 based phage. λZAP contains all the λ DNA sequences required for lytic growth, and in between these is the DNA sequence needed for plasmid replication and selection (the ColE1 ori and AMPR). Additionally, λZAP contains the lacZ gene and multiple cloning site sequence in a similar fashion to the pUC plasmids. The plasmid sequences in the vector begin with the f1 initiator and end with the f1 terminator. Vectors bearing foreign DNA can be selected by blue –white screening of λ plaques, and the insert DNA can be isolated in the form of a plasmid when bacteria harbouring the λ phage are superinfected with an f1 helper phage. Proteins produced by the helper phage will result in DNA replication between the f1 initiator and terminator. The
142 |
VECTORS |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
SacI NotI XbaI SpeI EcoRIXhoI |
|
|
|
|
|||
|
lZAPII |
|
|
|
|
|
MCS |
|
|
|
|
|||
|
cos |
|
|
|
|
|
|
|
cos |
|||||
|
|
AMPR |
ColE1 ori |
|
|
|||||||||
|
|
|
|
|
|
|
||||||||
|
|
Left l arm |
|
|
|
|
|
|
|
|
|
Right l arm |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
f1 |
|
|
|
|
lacZ′ |
f1 |
|
||||
|
|
|
terminator |
Phagemid DNA |
initiator |
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Construct DNA library |
|||||
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
Isolate positive clone |
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
cos |
|
|
|
|
Insert DNA |
|
|
|
cos |
||||
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Left l arm |
|
|
|
|
|
|
|
|
|
Right l arm |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
Excise plasmid containing |
|
||||
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
insert by co-infection with |
|||||
|
|
|
|
|
|
|
|
|
f1 helper phage |
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Initiator
Terminator
f1 ori
pBluescript
AMPR
lacZ′ InsertDNA
ColE1 ori
Figure 3.17. The in vivo excision of phagemid DNA from a λ phage vector. λZAPII is a sophisticated λ phage vector containing the elements of the λ and M13 phages as well as the sequences required for stable phagemid production. The DNA sequence for the entire pBluescript phagemid is contained within the λ vector between the f1 initiator and terminator. Foreign DNA inserted into the multiple cloning site (MCS) of λZAPII can be recovered in the form of a phagemid. Bacteria harbouring the λ phage are superinfected with an M13 based phage to drive DNA replication of sequences between the f1 initiator and terminator. The M13 phages produced using this DNA can be used to infect F E. coli cells and double-stranded plasmid DNA isolated
single-stranded DNA will circularize and will be packaged as an M13-like phage and secreted from the cell. The introduction of the M13 phage particles into an F E. coli strain and selection on ampicillin will result in the formation of colonies containing the recombinant plasmid, which can then be isolated as double-stranded plasmids.
3.7 ARTIFICIAL CHROMOSOMES |
143 |
|
|
3.7Artificial Chromosomes
The major limitation of most of the vectors that we have discussed so far is the size limit of the DNA that can be cloned into them. Natural eukaryotic chromosomes consist of hundreds or thousands of genes, together with DNA elements required for chromosomal stability and function such as telomeres and centromeres. Telomeres, which consist of DNA and protein, are located at the ends of chromosomes and protect them from damage. Centromeres are segments of highly repetitive DNA that are essential for the proper control of chromosome distribution during cell division. A logical extension of vector design to clone very large DNA fragments is, therefore, to reconstruct an autonomously replicating chromosome into which DNA fragments may be cloned. Cloning in this way is conceptually similar to cloning in λ phage – with the reconstruction of a replication competent DNA molecule – except that the scale of the foreign DNA that can be cloned is much greater.
3.7.1YACs
Yeast artificial chromosome (YAC) vectors allow the cloning, within yeast cells, of fragments of foreign genomic DNA that can approach 500 kbp in size. These vectors contain several elements of typical yeast chromosomes, including the following.
•A yeast centromere (CEN4). The yeast centromere is specified by a 125 bp DNA segment. The consensus sequence consists of three elements: a 78 –86 bp region with more than 90 per cent AT residues, flanked by a conserved sequence on one side and a short consensus sequence on the other (reviewed by Clarke (1990)).
•Yeast autonomously replicating sequence (ARS1). Yeast ARS elements are essentially origins of replication that function in yeast cells autonomously from the replication of yeast chromosomal replication origins.
•Yeast telomeres (TEL). Telomeres are the specific sequences (5 - TGTGGGTGTGGTG-3 ) that are present at the ends of chromosomes in multiple copies and are necessary for replication and chromosome maintenance.
•Genes for YAC selection in yeast. The vector has a functional copy of URA3, a gene involved in uracil biosynthesis, and TRP1, a gene involved in tryptophan biosynthesis, that allow selection of yeast cells that have taken up the vector. The YAC is transformed into a host yeast cell that is defective in these biosynthetic pathways, and transformants are identified by their ability to complement the nutritional defect.
144 |
VECTORS 3 |
|
|
•Bacterial replication origin and a bacterial selectable marker. In order to propagate the YAC vector in bacterial cells, prior to insertion of genomic DNA, YAC vectors usually contain the ColE1 ori and the ampicillin resistance gene for growth and analysis in E. coli.
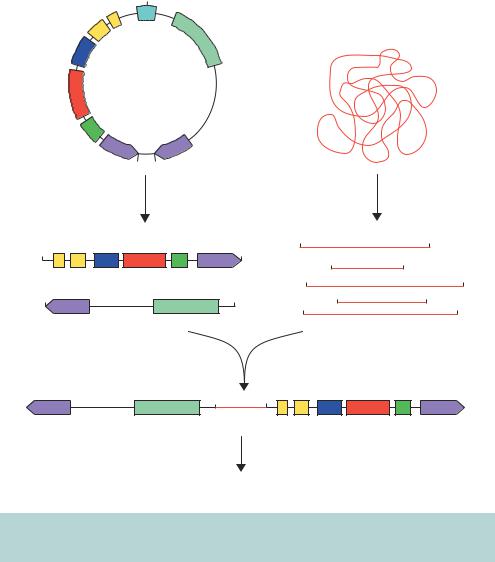
The cloning of DNA fragments into a YAC is shown diagrammatically in Figure 3.18. The YAC is cleaved using restriction enzymes to generate two ‘arms’ that each have a telomere sequence at the end. One of the arms contains an autonomous replication sequence (ARS1), a centromere (CEN4) and a
|
|
EcoRI |
|
|
|
|
|
||
|
CEN4 |
|
|
|
|
|
|
|
|
|
ARS 2 |
SUP4 |
URA3 |
|
|
|
|||
|
|
|
Genomic DNA |
||||||
TRP1 |
|
|
|
|
|
||||
|
|
|
|
|
|
|
|||
AMPR |
pYAC4 |
|
|
|
|
|
|||
|
|
|
|
|
|
|
|||
|
ori |
TEL |
TEL |
|
|
|
|
|
|
|
BamHI |
BamHI |
|
|
|
|
|
||
|
|
|
Cut with EcoRI |
|
|
|
Partial digest |
||
|
|
|
and BamHI |
|
|
|
with EcoRI |
||
EcoRI |
ARS2 |
AMPR ori |
TEL |
|
|
EcoRI |
EcoRI |
||
BamHI |
EcoRI |
EcoRI |
|||||||
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|||
CEN4 TRP1 |
|
|
|
|
|
EcoRI |
EcoRI |
||
BamHI |
TEL |
|
URA3 |
EcoRI |
|
EcoRI |
EcoRI |
||
|
|
|
|
|
|
|
EcoRI |
EcoRI |
|
|
|
|
|
|
|
Mix and ligate |
|
||
TEL |
|
URA3 |
EcoRI EcoRI |
ARS2 |
AMPR ori TEL |
||||
|
|
|
|
|
|
||||
|
|
|
|
Genomic |
|
|
|||
|
|
|
|
|
DNA |
CEN4 TRP1 |
|
||
Transform into ade−, ura−, trp− yeast and select for red, URA+, TRP + colonies
Figure 3.18. Cloning of very large DNA fragments into a YAC vector. See the text for details
3.7 ARTIFICIAL CHROMOSOMES |
145 |
|
|
selectable marker (TRP1). The other arm contains a second selectable marker (URA3). Large DNA fragments (>100 kbp) are then ligated between the two arms (Anand, Villasante and Tyler-Smitu, 1989). The insertion of foreign DNA into the cloning site inactivates the suppressor tRNA gene SUP4, expressing tRNATyr, in the vector DNA. In an ade2–ochre host yeast cell, the expression of SUP4 results in the formation of white colonies, while in those in which it has been insertionally inactivated will give rise to red yeast colonies (Burke, Carle and Olson, 1987). Yeast cells that are mutated in the ADE2 gene product (coding for the enzyme phosphoribosylamino-imidazole-carboxylase) have a block in the adenine biosynthetic pathway, causing an intermediate to accumulate in the vacuole. This intermediate gives the cell a red colour. The recombinant YACs are therefore transformed into a yeast strain that has defects in its chromosomal copies of the ura3, trp1 and ade2 genes. Transformants are identified as those red colonies that grow on media lacking both uracil and tryptophan. This ensures that the cell has received an artificial chromosome with both telomeres (because of complementation of the two nutritional mutations) and the artificial chromosome contains insert DNA (because the cell is red).
There are difficulties associated with working with YACs. Some of these are listed below.
•Very large DNA molecules are very fragile and prone to breakage, leading to problems of rearrangement.
•It is estimated that between 10 and 60 per cent of clones in YAC genomic libraries are chimaeric, i.e. regions from different parts of the genome become joined in a single YAC clone (Green et al., 1991).
•Clones tend to be unstable, with their foreign DNA inserts often being deleted. Naturally occurring repetitive DNA sequences are rare in the yeast genome, and the insertion of such sequences from, say, human DNA inserts appears to increase the recombination frequency within the YAC. This may make the YAC unstable. Interestingly, however, larger YAC vectors are more stable in yeast than shorter ones, which consequently favours cloning of large stretches of DNA (Smith, Smyth and Moir, 1990).
•There is a high rate of loss of the entire YAC during mitotic growth.
•It is difficult to separate the YAC from the other host chromosomes because of their similar size. Separation requires sophisticated pulsed-field gel electrophoresis (PFGE).
•The yield of DNA is not high when the YAC is isolated from yeast cells.
146 |
VECTORS 3 |
|
|
3.7.2PACs
To overcome some of the problems associated with using cosmid or YAC systems, a method for cloning and packaging DNA fragments using a bacteriophage P1 system has been developed that offers the ability to clone large genomic DNA fragments of between 70 and 95 kbp in size. P1 bacteriophage has a much larger genome than λ phage (in the range of 110 –115 kbp), and vectors have been designed with the essential replication components of P1 incorporated into a plasmid (Ioannou et al., 1994). Upon infecting E. coli, bacteriophage P1 may either express its lytic functions, producing 100 –200 new bacteriophage particles and lysing the infected bacterium, or the infecting bacteriophage may repress its lytic functions, and the bacteriophage genome is maintained as a large, stable, low-copy plasmid. P1 phage has two replication origins – one to control lytic DNA replication, and the other to maintain the plasmid during non-lytic growth. During the lytic cycle, new phage DNA is produced and cleaved at a pac site prior to insertion into phage particles.
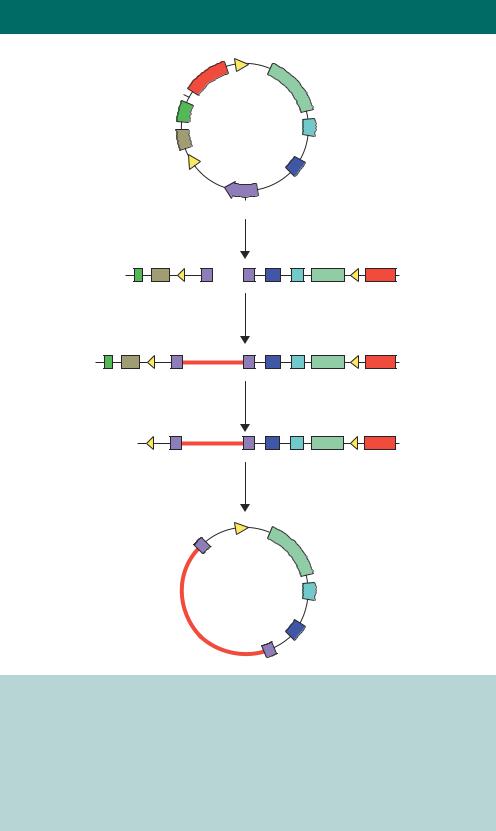
The cloning of foreign DNA fragments into a P1 vector, or P1 artificial chromosome (PAC), is shown in Figure 3.19. The PAC vector is digested with the restriction enzymes ScaI and BamHI to generate two vector arms: a short
and a long arm. Genomic DNA is partially digested with MboI (recognition sequence 5 -GTAC-3 , yielding BamHI-compatible sticky ends) and size selected on a sucrose gradient. Fragments between 70 and 95 kb in length are isolated and ligated in between the vector arms to generate a series of linear molecules. If ligation occurs between two short arms, the resulting molecule will contain neither the P1 replication origins nor the KANR gene, and will be non-viable. If both arms are long there will be no pac site, and no packaging into the phage heads will occur. The only viable recombinant will consist of the insert sequence flanked by both a short and long arm. Phage P1 uses a ‘head-full’ packaging strategy and can accommodate a total DNA length of approximately 110 –115 kbp. This means that any inserts longer than 95 –100 kbp will result in the truncation of the packaged DNA before both loxP sites are inserted into the
phage, and the molecule will be unable to circularize upon transfection into the host. Once injected into the cre+ E. coli host cell, the Cre protein circularizes the DNA at the loxP sites, and DNA then replicates using the plasmid origin of
replication. The original vector BamHI restriction enzyme site, into which the foreign DNA was inserted, is located within the bacterial sacB gene (encoding levansucrase). The expression of this gene is toxic to E. coli cells growing on sucrose. Thus sucrose growth provides a mechanism of positive selection for those PACs containing inserts. Propagation of E. coli cells harbouring the recombinant PAC on media containing sucrose permits growth of colonies with DNA inserts.
|
|
|
|
3.7 |
ARTIFICIAL CHROMOSOMES |
147 |
|
|
AMPR |
loxP |
P1 plasmid |
|
|||
|
|
|
|
||||
|
|
|
replicon |
|
|||
|
ScaI |
|
|
|
|
||
|
|
|
|
|
|
|
|
|
ori |
|
|
|
|
|
|
|
|
|
PAC vector |
KANR |
|
||
|
pac |
|
|
|
|
|
|
|
|
|
loxP |
|
|
|
|
|
|
|
|
sac B |
P1 lytic |
|
|
|
|
|
|
|
replicon |
|
|
|
|
|
BamHI |
|
|
|
|
|
|
|
|
BamHI and ScaI |
|
|
|
ori |
pac loxP |
|
KANR |
loxP AMPR |
|
||
|
|
|
+ |
|
|
|
|
|
|
|
|
P1 lytic |
P1 plasmid |
|
|
|
|
|
|
replicon |
replicon |
|
|
|
|
|
|
Ligate genomic DNA |
|
|
|
ori pac |
loxP |
70–95 kbp |
KANR |
loxP AMPR |
|
||
|
|
|
|
|
|
||
|
|
|
|
P1 lytic |
P1 plasmid |
|
|
|
|
|
|
replicon |
replicon |
|
|
|
|
|
|
Package in vitro |
|
|
|
|
loxP |
|
|
KANR |
loxP AMPR |
|
|
|
|
|
|
P1 lytic |
P1 plasmid |
|
|
|
|
|
|
replicon |
replicon |
|
|
|
|
|
|
Transfect cre+ E. coli cells |
|
||
|
|
|
|
loxP |
P1 plasmid |
|
|
|
|
|
|
|
replicon |
|
|
|
|
|
Recombinant |
KANR |
|
||
|
|
|
|
PAC |
|
|
|
P1 lytic replicon
Figure 3.19. Cloning into a PAC vector. The PAC vector contains the P1 bacteriophage plasmid and lytic replicons, together with a pac cleavage site to allow DNA assembly into phage particles. Additionally, the vector contains the pUC origin of replication (ori) and ampicillin resistance gene for the propagation and selection of the vector itself. These sequences are lost in the recombinant PAC. Large DNA inserts are cloned into the sacB gene (whose function can be selected against) and packaged in vitro into P1 phage particles. Transfection of the P1 phage particles into an E. coli cell harbouring a copy of the Cre recombinase will result in circularization of the recombinant PAC at the loxP sites. The circular form is then maintained at low copy number using the P1 plasmid replicon
