
John Wiley & Sons - 2004 - Analysis of Genes and Genomes
.pdf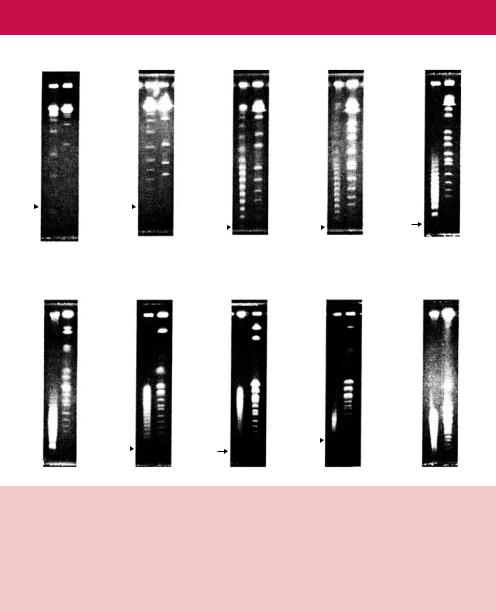
98 |
|
BASIC TECHNIQUES IN GENE ANALYSIS |
2 |
|
|
|||||
|
|
15 sec |
|
30 sec |
|
|
45 sec |
|
|
1 min |
|
|
– 273 |
|
|
– 440 |
– 677 |
|
– 836 |
||
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
– 600 |
|
– 792 |
|
|
– 214 |
|
|
– 341 |
|
– 752 |
|||
|
|
|
|
– 551 |
|
|||||
|
|
|
|
|
– 667 |
|||||
|
|
|
|
|
– 273 |
|
|
|||
|
|
|
|
|
– 440 |
|
|
|||
|
|
|
|
|
– 214 |
|
|
|||
|
|
|
|
|
|
|
|
|||
48.5 |
|
|
|
|
|
|
|
– 214 |
|
– 214 |
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
1.5 min |
|
1.75 min |
2 min |
|
2.5 min |
|
|
|
– 2200 |
|
– 2200 |
– 1125 |
|
|
– 1600 |
|
– 1600 |
|
|
|
|
||
|
|
|
|
|
|
– 960 |
|
– 1125 |
– 1125 |
|
|
|
|
|
|
|
|
|
|
|
|
|
– 214 |
– 214 |
|
– 214 |
– 214 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
48.5 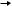
1.25min
–960
–914
–836
–214
3 min
– 2200
– 1600
– 214
Figure 2.19. The effect of pulse time on the separation of DNA fragments of different sizes. The chromosomes of the yeast Saccharomyces cerevisiae and a set of known molecular size DNA markers were subjected to PFGE under otherwise identical conditions using different pulse times. Notice that increasing the pulse time gives rise to better separation of large DNA fragments. The sizes of the markers are shown in kbp. Reproduced from Wrestler et al. (1996) by permission of Bruce Birren (Whitehead Institute)
bacteria or yeast) in agarose plugs and then treat the plugs with enzymes to digest away the cell wall and proteins, thus leaving the naked DNA undamaged in the agarose. The plugs then are cut to size, treated with restriction enzymes if necessary and then loaded into the well of an agarose gel (Figure 2.20).
2.6Nucleic Acid Blotting
When we look at DNA fragments on an ethidium bromide stained gel, all the DNA molecules appear to be identical (Figure 2.13(a)). We know, however,
|
2.6 NUCLEIC ACID BLOTTING |
99 |
|
|
|
|
Molten |
|
Intact cell |
agarose |
|
Mix and pour into block former. Allow to cool
Incubate with hydrolytic enzymes
Digestion of cells
DNA trapped
Agarose blocks of DNA loaded into agarose gel slots
Figure 2.20. Preparation of high-molecular-weight DNA for analysis by PFGE. Intact cells are mixed with molten, but cool, agarose and poured into a block former. Once set, the agarose blocks can be treated with hydrolytic enzymes to break open the cell walls and release the chromosomal DNA. If required, the DNA can be digested with restriction enzymes whilst also in the agarose block. The treated block is then loaded into the well of an agarose gel before being subjected to PFGE
that the sequence of the DNA in each band on the gel is different to other bands – even bands that are identical in length may have a very different DNA sequence. It is, of course, the DNA sequence itself that plays a vital role in the function of a molecule. In the mid-1970s methods were developed to distinguish
100 |
BASIC TECHNIQUES IN GENE ANALYSIS 2 |
|
|
bands on gels that contain a particular DNA sequence. These methods rely on the hybridization of nucleic acid sequences in order to detect the presence of complementary sequences. The original method of blotting was developed by Ed Southern in 1975 for detecting DNA fragments in an agarose gel that were complementary to a given nucleic acid sequence (Southern, 1975).
2.6.1Southern Blotting
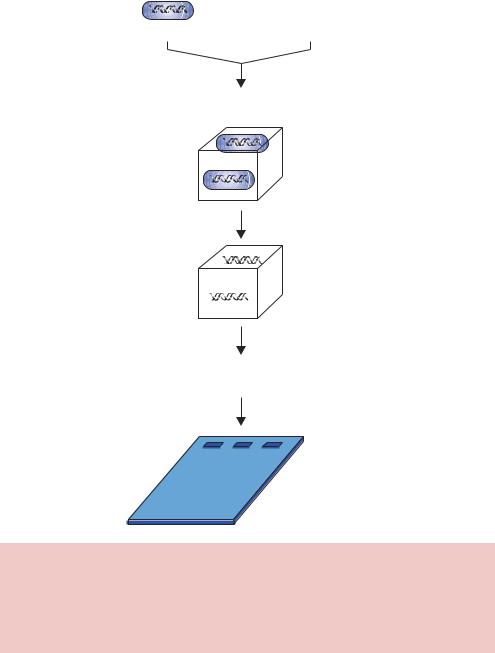
In the procedure, referred to as Southern blotting, the DNA fragments separated on an agarose gel are transferred and immobilized onto a membrane. After the gel has been run, the DNA fragments are denatured (i.e. the strands are separated) using alkali. The single-stranded nature of the DNA on the membrane is important to allow complementary DNA sequences to be able to bind to the DNA fragments attached to the membrane. After the DNA in the gel has been denatured, the DNA fragments are transferred to a membrane. The transfer process can be mediated either electrophoretically or through capillary attraction by placing the gel and membrane in contact with each other and allowing buffer to flow through the gel onto the membrane – the DNA fragments move with the buffer and become trapped on the membrane. Initially, nitrocellulose membranes were used, but these were fragile and easily broken. Nylon membranes are commonly used today. After transfer, the DNA
fragments need to be fixed to the membrane so that they cannot detach. A number of methods of fixing are available including baking at 80 ◦C and ultraviolet cross-linking. UV cross-linking is based on the formation of crosslinks between a small fraction of the T residues in the DNA and the positively charged and amino groups on the surface of the nylon membrane. Following fixation, the membrane is placed in a solution of labelled (often radioactive) single-stranded nucleic acid – either single-stranded DNA or RNA. The labelled nucleic acid (or probe) is allowed to hybridize to its complementary partner sequence on the membrane. The interaction between the single-stranded probe and its complementary sequence will result in the binding of the probe to the membrane through non-covalent hydrogen bonding that normally holds the DNA double helix together. The membrane is then washed extensively to remove non-specifically bound probe, and specific interactions are detected by exposing the membrane to X-ray film (Figure 2.21).
Southern blotting is used to detect DNA sequences that are either identical or similar to the sequence of the probe. The hybridization of the probe to the DNA sequences trapped on the membrane is the critical component of success of these experiments. As we have seen previously, single-stranded DNA will bind with high affinity to its complementary partner sequence. It will also

2.6 NUCLEIC ACID BLOTTING |
101 |
|
|
DNA separated through agarose gel
Blot DNA onto membrane
Add single-stranded labelled DNA
Expose membrane to X-ray film
Band on film shows position of complementary DNA sequences
Figure 2.21. Southern blotting. DNA fragments separated on an agarose gel are transferred onto a membrane. The DNA is made single stranded before the addition of a labelled probe. The binding of the probe to the membrane – through base pair hydrogen bonding – can be detected by exposing the membrane to X-ray film. The radio-labelled probe will form a band on the film at a position corresponding to the complementary sequence on the membrane
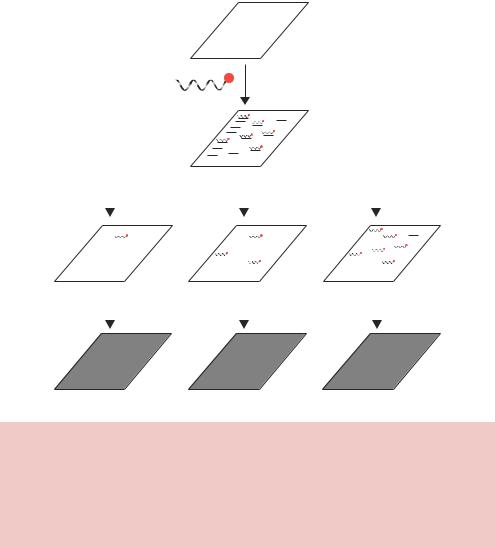
bind to similar, but non-identical, DNA sequences with a reduced affinity. This differential binding affinity to different DNA sequences can be used to identify DNA molecules that are not identical but are merely related to the probe. This sort of analysis is achieved by altering the temperature (or salt concentration) at which the probe is washed off the filters after it has bound (Figure 2.22). Washing the membrane at high temperature (high stringency) will result in thermal disruption of all but the most tightly bound sequences. Consequently, the bands that show up on the X-ray will film will be either identical or highly related to the probe. Washing the membrane to lower temperatures will reduced the overall level of stringency, and sequences that are less related to the probe
102 |
BASIC TECHNIQUES IN GENE ANALYSIS 2 |
||||||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Add single-stranded labelled DNA
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
65°C |
|
50°C |
|
|
|
|
|
|
|
|
|
|
35°C |
|
|
|
|
|
|
||||||||||||||||||||||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
High strigency |
Medium strigency |
Low strigency |
Figure 2.22. Washing Southern blot membranes at different temperatures results in different stringencies. Washing the membrane at high temperature will remove all but the most tightly bound DNA molecules – those most similar to the sequence of the probe. Lower-temperature washes can reveal sequences on the membrane that are similar, but not identical, to the sequence of the probe. The actual temperature at which washes are performed will depend on the length of the probe and the likelihood of it finding an exactly matching sequence on the membrane
will give positive signals on the X-ray film. This approach is immensely useful when you do not precisely know the sequence of the gene you wish to identify. This situation often arises when knowledge of the protein sequence is known, but the DNA sequence encoding that protein is unknown. We shall discuss this issue further in Chapter 6.
2.6.2The Compass Points of Blotting
Since Southern’s first description of blotting and hybridization, a number of variants have been described. These all involve the transfer of material from a
2.7 DNA PURIFICATION |
103 |
|
|
gel to a membrane, and subsequent specific detection of particular molecules. Such techniques have been assigned names reflecting the points of a compass rather than a description.
•Northern blotting – the detection of RNA sequences using probes (Thomas, 1980). RNA is separated on an agarose gel and transferred to a membrane, much as described above. Specific RNA molecules are then detected by hybridization using labelled single-stranded DNA or RNA sequences that are complementary to particular RNAs.
•Western blotting – the detection of specific proteins using antibodies (Burnette, 1981). Proteins are separated through a polyacrylamide gel containing the detergent SDS to keep them in an unfolded (denatured) state. The proteins are transferred from the gel onto a membrane in much the same way as described above for Southern blotting. Particular proteins are then detected using antibodies. The specific interaction between the antibody and its antigen occurs on the membrane, and the position of the bound antibody is detected. Initially radio-labelled antibodies were used, but these have been largely superseded by antibody ‘sandwiches’. The sandwiches work through the binding of one unlabelled antibody (the primary antibody) to the antigen on the membrane. A second, labelled, antibody (the secondary antibody) is then used to detect the presence of the first antibody. This has several advantages; firstly, the multivalent nature of antibody binding means that a substantial increase in sensitivity is achieved, and secondly, a single secondary antibody can be used to detect a number of different primary antibodies. For example, primary monoclonal antibodies are often raised in mice. A secondary antibody raised, say, in the rabbit against mouse immunoglobulins will be capable of interacting with a number of different mouse derived primary antibodies (Figure 2.23).
•South-Western blotting – the identification of proteins that bind to particular DNA sequences (Singh et al., 1988). Proteins are either separated on a gel or blotted directly onto membranes. The membrane is then challenged with a labelled double-stranded DNA oligonucleotide. If a protein is able to bind the DNA, it will be detected by the presence of the label.
2.7DNA Purification
Many of the techniques we have discussed in this chapter are heavily reliant upon the availability of relatively large quantities of purified DNA. The isolation of nuclear material from cells is a relatively straightforward process. Lysis of
104 |
BASIC TECHNIQUES IN GENE ANALYSIS 2 |
||||||||||||||||||||||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Proteins separated through polyacrylamide gel
Blot proteins onto membrane
Block non-specific protein binding sites and add primary antibody
Antibody binds to antigen
Wash away excess primary and add labelled secondary antibody
Antibody sandwich
Detect label
Band shows position of antigen
Figure 2.23. Western blotting using antibodies to detect specific proteins. Proteins are separated through a polyacrylamide gel, usually containing SDS to maintain them in a denatured state, prior to blotting onto a nitrocellulose membrane. Non-specific protein binding sites are blocked on the membrane – using solubilized milk powder – before the primary antibody is added. The primary antibody will specifically bind to the antigen to which it was raised. A labelled secondary antibody is then added to detect the location of the primary antibody. The antibody sandwich results in an amplification of the signal. The secondary antibody is often labelled with an enzyme whose activity, in the presence of appropriate substrates, results in either a colour change on the membrane or the emission of light that can be detected using X-ray film
the cell wall will result in the nuclear material spilling out from the broken cells. This material can be harvested, but the DNA will be contaminated with both RNA and with proteins. Methods are therefore required to remove these contaminants to yield a purified DNA fraction.
2.7 DNA PURIFICATION |
105 |
|
|
Total genomic DNA isolation is often carried out according to the following procedure.
•The host cell wall is lysed. The method used for the lysis procedure depends upon the nature of the host cell itself. For instance, bacterial cells are often treated with the enzyme lysozyme to weaken their cell wall before being lysed with detergents. Yeast cells, on the other hand, are treated with zymolase to disrupt the integrity of their cell wall before lysis proceeds – often by grinding the cells using glass beads to break them open.
•The cellular debris will be removed by centrifugation.
•The remaining soluble material will then be vortexed in the presence of phenol to remove proteins by denaturing then. Chloroform is often used in conjunction with phenol (as a phenol/chloroform solution) since it is also a protein denaturant, but it also stabilizes the rather unstable boundary between the aqueous phase and a pure phenol layer.
•The nucleic acid remaining in the aqueous layer can then be precipitated using ethanol. Upon addition of ethanol the DNA will form a stringy white precipitate that can be collected by a centrifugation.
•RNA can be removed from the preparation by treatment with RNase.
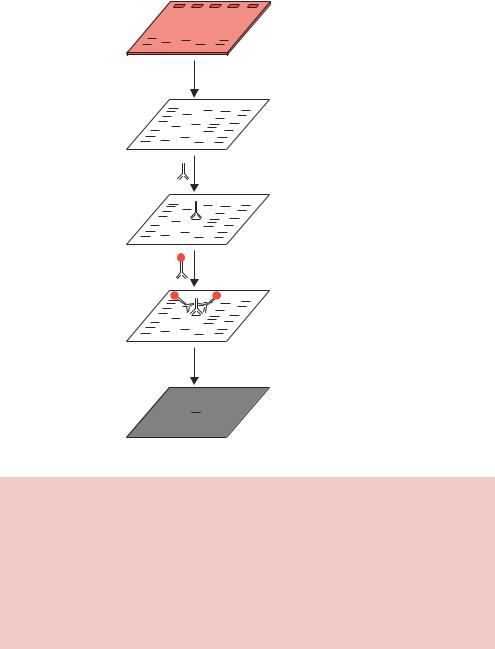
For many applications, the DNA must be purified further. Isopycnic centrifugation can be used to separate DNA from contaminants, and also to isolate individual types of DNA molecule. The DNA sample is mixed with a substance (caesium chloride, CsCl) that is capable of forming a self-generating uniform density gradient during the centrifugation process. DNA molecules will sediment only to the position in the centrifuge tube at which the gradient density is equal to its own density, and they will remain there to form a band. After the DNA band has formed, it is removed from the centrifuge tube and the CsCl removed by precipitating the DNA with alcohol. A major advantage of this technique is the ability to separate different species of DNA molecules (Figure 2.24). The inclusion of ethidium bromide in the centrifugation mix results in its binding to DNA. The topological constraints of supercoiled DNA mean that closed-circular DNA molecules (e.g. plasmids) are able to bind only limited amounts of ethidium bromide. Nicked DNA circles, and linear DNA fragments, are able to bind much more ethidium bromide and are extensively unwound in its presence (Figure 2.12). This results in nicked and linear DNA being less dense than their supercoiled equivalent, and consequently being separated in a different location on a CsCl/EtBr gradient. The main drawback of isopycnic centrifugation is the long hours of centrifugation required to form

106 |
BASIC TECHNIQUES IN GENE ANALYSIS 2 |
|
|
density CsCl
Protein

 Linear DNA
Linear DNA

 Nicked DNA
Nicked DNA
Supercoiled DNA |
RNA |
Figure 2.24. Isopycnic centrifugation of DNA molecules through a caesium chlo- ride–ethidium bromide gradient. The DNA sample is mixed with caesium chloride and ethidium bromide and centrifuged at high speed for approximately 48 h. Each component bands at it own density, which, for the DNA, is influenced by the extent to which the molecule can be unwound by the ethidium bromide
the gradients, with isopycnic DNA banding taking 36 –48 h to form in a CsCl gradient. The technique has lost favour recently with the introduction of commercially available ‘kits’ that allow rapid and inexpensive DNA isolation with minimal exposure to harmful reagents. The basis of these ‘kits’ is discussed below.
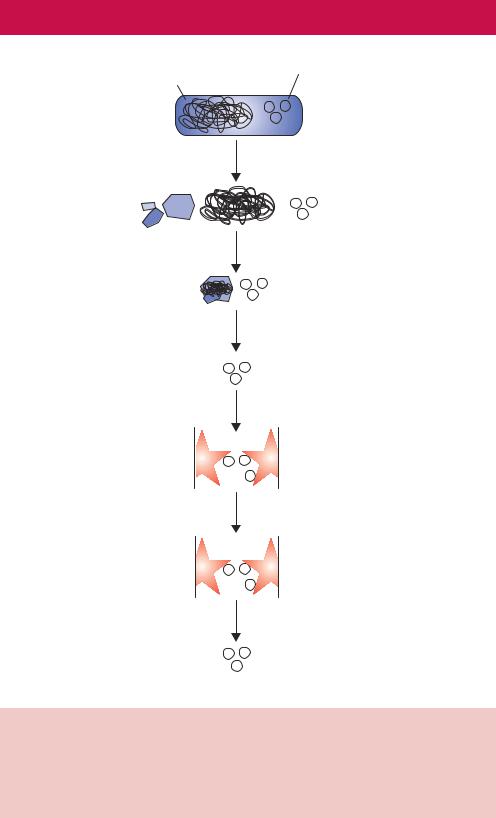
The purification of plasmid DNAs (see Chapter 3) is vitally important for cloning and the in vitro manipulation of genes. An obvious problem is distinguishing the relatively small amount of plasmid DNA contained within a cell from the relatively large amount of chromosomal DNA that will also be present. Plasmid DNA is most commonly isolated from bacterial cells. Again, cell lysis is an important stage in the process. Insufficient cell lysis will result in low plasmid yields, while cells that have been lysed too much suffer similar problems. The most popular methods of extracting DNA from cells are based on the original procedure of Birnboim and Doly (1979). This method, termed the alkaline lysis procedure, takes advantage of the fact that at alkali pH (between 12.0 and 12.5) linear DNA, but not closed-circular DNA, will become denatured. These procedures inevitably result in shearing of the circular E. coli genome to generate large linear DNA fragments. Most methods to purify plasmid DNA based on this observation require small cultures of
|
2.7 DNA PURIFICATION |
107 |
High molecular weight |
Plasmid DNA |
|
genomic DNA |
|
|
Bacterial cell
SDS + sodium hydroxide
Neutralise with potassium acetate
Genomic DNA  and proteins precipitated
and proteins precipitated
Remove precipitate by centrifugation
Plasmid DNA in solution
Bind to gel matrix
Elute impurities
Elute DNA
Pure plasmid DNA
Figure 2.25. Plasmid DNA purification. Bacterial cells harbouring plasmid DNA are broken open using SDS. The presence of sodium hydroxide ensures the denaturation of genomic DNA. Upon neutralization, the genomic DNA aggregates and forms a precipitate together with the SDS–protein complexes. These can be removed by centrifugation. The plasmid DNA is then bound to a matrix to which contaminants are unable to adhere. The pure plasmid DNA can then be eluted from the matrix
