
Neutron Scattering in Biology - Fitter Gutberlet and Katsaras
.pdf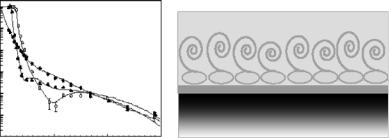
14 Biomimetic Structures at Surfaces and Interfaces |
295 |
Adjacent to the surface, a dense protein layer was observed that protrudes into the solution with a structure of lower density as schematically shown in Fig. 14.5. A proteolytic enzyme, endoproteinase Asp-N, which cleaves the hydrophilic part of β-casein, a ected only the outermost β-casein layer. Similar experiments were performed with β-lactoglobulin [62]. E ects of surfactants and of cations on β-casein adsorption to hydrophobized silica were also investigated [63, 64]. In comparison, protein adsorption to bare (i.e., unmodified), hydrophilic silica interfaces showed similar structural features, albeit at significantly slower adsorption rates [65]. The study thus demonstrates well that neutron reflection may be useful for studies of dynamics of interfacial phenomena in protein systems.
In a similar vein, the adsorption of lysozyme to OTS modified interfaces was observed to be irreversible [67]. The protein formed a densely packed, thin layer at the OTS surface with a di use, thicker proportion facing the bulk solution – a similar structure as observed with β-casein. As none of the dimensions of these structures corresponded to those of the globular protein in solution
– unlike after adsorption at the hydrophilic silica–water interface – lysozyme is believed to denature at the OTS–water interface. The interfacial structure of lung surfactant has been studied at OTS-hydrophobized silica solid–liquid interfaces [68]. Results on lung surfactant from rabbit were interpreted within a two-layer model. An inner layer of 20 ˚A thickness contained about 50% solvent, consistent with a loosely packed phospholipid monolayer with substantial amounts of solvent and protein incorporated. The outer layer was 80 ˚A thick and incorporated 10% protein. Addition of the water-soluble phospholipase PLA2 led to a densification of both layers. Whether these findings bear implications for a general understanding of surfactant function remains to be seen, since porcine lung surfactant showed somewhat di erent behavior. A phospholipid monolayer of higher density was observed, and the addition of PLA2 exerted much less e ect than in the rabbit system.
(a)
Reflectivity
1
10−1
10−2
10−3
10−4
10−5
10−60
(b)
0.05 |
0.1 |
0.15 |
qz(Å−1)
Water
β-Casein
OTS
Silicon
Fig. 14.5. (a) Neutron reflectivity and model fits measured for β-casein adsorbed at pH 7 on deuterated OTS in (◦) D2O, (•) H2O, and ( ) water with an SLD of 4.5 × 10−6 ˚A−2 (details see [61]). (b) Schematic interpretation of these results [3]
296 T. Gutberlet et al.
14.5.2 Hydrophilic Modified Interfaces
Self-assembled monolayers (SAMs) provide a well-established method for the control of the surface-chemistry of solid substrates. Neutron reflectivity is among the standard techniques for the molecular-scale characterization of such interfaces. It has, for example, been used to characterize the adsorption of human serum albumin (HSA) onto silicon-supported NH+3 -terminated SAMs [69]. Upon incubation with high protein concentration (0.1% wtV−1), HSA formed a two-layer adsorption structure. Directly adjacent to the solid surface a 40 ˚A thick protein layer was observed. A secondary layer extended an additional 40 ˚A into the solution.
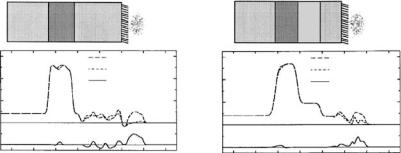
In a slightly di erent context, chemical interface modification with SAMs also enables stable anchoring of biomolecules to polar or apolar surfaces [70]. For example, yeast cytochrome c (YCC) was covalently bound to the interface of a SAM with mixed –CH3/–SH or –OH/–SH endgroups via disulfide linkage between a cysteine residue on the protein surface and the exposed thiol moieties of the SAM. If the SAM is prepared on top a stratified Fe/Si or Fe/Au/Si surface nanostructure, the resulting SLD striations allow for an interferometric approach to data inversion. This approach reduces the intrinsic limitation of the resolution that is due to the restrictions in attainable Qz range. The potential of similar approaches for an unambiguous phase determination to calculate the SLD profile [71] is described in detail in the contribution by Majkrzak et al., in this volume. Measurements of the system in air with H2O and D2O hydrating the protein monolayer provided water distribution profiles [70] (Fig. 14.6). These profiles were consistent with corresponding electron density profiles determined previously via X-ray interferometry [72].
As discussed above, biotin–streptavidin technology is another standard that enables biospecific ligation at interfaces. In the context of functionalization of solid interfaces, streptavidin has been immobilized at an aminopropylterminated silicon surface. Biotinylated DNA strands have subsequently been grafted to this surface [73], and the evolution of the complex structure has been followed by neutron reflectometry. It was determined that DNA is collapsed on the surface of the streptavidin layer due to its electrostatic interaction with the positively charged streptavidin. Similarly, the formation of a lignin layer immobilized on a SAM-terminated Si surface has been investigated [74]. The lignin film was grown on an aminopropyl modified Si-wafer by grafting of this surface with polysaccharides and peroxidase, followed by polymerization at the solid–liquid interface. Intermediate and final structures were studied by neutron reflectometry using H2O–D2O contrast variation.
PEG-coated surfaces have been shown to prevent protein adsorption. Using neutron reflectivity, this was demonstrated for PEGs with a molecular weight of ≈5000 Da [75] as well as for short-chain SAMs of methoxy-tri(ethylene glycol) [76, 77]. A di erent way to passivate the hydrophilic silicon oxide surface against protein adsorption is by chemically anchoring an organic monolayer bearing terminal phosphorylcholine (PC) groups, as described in the
14 |
Biomimetic Structures at Surfaces and Interfaces |
297 |
(a) |
(b) |
|
] −2 Å −6 [x10b
r
15 |
D2O |
10 |
H2O |
Difference |
|
|
(offset by 5 unit) |
5 |
|
0 |
|
−5 |
|
−250 −200 −150 −100 −50 |
0 50 100 |
Z [Å]
] −2 Å −6 [x10b
r
15 |
D2O |
10 |
H2O |
Difference |
(offset by 5 unit)
5
0
−5
−250 −200 −150 −100 −50 0 50 100
Z [Å]
Fig. 14.6. Neutron SLD profiles of YCC on SAMs with probe spins parallel to the iron magnetization for partial hydration with D2O and H2O, and their di erence profile for both a nonpolar (a) and a polar, (b) SAM. Schematic representations of the composite structures are shown on top [70]
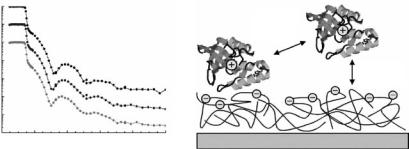
contribution by Lu in this volume. The approach was shown to be e ective in reducing protein adsorption of lysozyme, fibrinogen, and bovine serum albumin (BSA) [78]. Salt-induced protein resistance of poly(acrylic acid) (PAA) brushes against BSA has been demonstrated by neutron reflectometry [79]. BSA molecules penetrates deeply into the PAA brush at low sodium chloride concentrations, but did not interact with the PE cushion at >500 mM salt. The adsorption of enzyme staphylococcal nuclease (SNase) on negatively charged poly(styrene sulfonate) surface has been studied with neutron reflectometry [80] and it was observed that the degree of protein adsorption onto the charged surface depends largely on direct protein–protein interactions (Fig. 14.7).
14.6 Functionalized Lipid Interfaces
and Supported Lipid Bilayers
14.6.1 Solid-Supported Phospholipid Bilayers
Phospholipid bilayers attached to solid supports are considered model systems that may be useful for the investigation of biological membranes of limited complexity, thus hoped to o er insight into underlying organization and interaction principles. Preparation of such systems can be performed either by spontaneous fusion of vesicles onto solid–liquid interfaces [81] or by Langmuir– Blodgett transfer techniques [82]. Neutron reflectometry is one of the work horses applied to probe such systems at the molecular level. For example, the structure of DMPC vesicles adsorbed on planar quartz surfaces [83] and that of DPPC on silicon single crystals [84] has been investigated. The formation of single phospholipid bilayers immobilized on solid surfaces was thus demonstrated and found to be separated from the surface by a water layer
298 T. Gutberlet et al.
neutron reflectivity
100
10−1
10−2
10−3
10−4
10−5
10−6
10−7
0.00 0.02 0.04 0.06 0.08 0.10 Q/Å−1
proteinprotein interaction
proteinsurface interaction
Fig. 14.7. Left: Neutron reflectivity curves of silica–water interface coated with a negatively charged polyelectrolyte. The upper curve refers to this interface without adsorbed protein, the other curves have been measured when staphylococcal nuclease was adsorbed (middle: 23◦C, lower : 43◦C). Right: Sketch of positively charged staphylococcal nuclease adsorbed on negatively charged polyelectrolyte surface. The degree of adsorption largely depends on protein–protein interactions, whereas attractive electrostatic interactions between the protein and the surface play a minor role [80].
of a few ˚Angstrom thickness. The process of phospholipid bilayer formation has been followed by time resolved measurements [85] (Fig. 14.8). Results were consistent with AFM and quartz microbalance (QMB) studies [86–88]. The interaction of biomolecules with such supported lipid bilayers has been exploited in various studies. For example, the penetration of the bacterial toxin, pneumolysin into mixed phospholipid membranes (10:10:1 molar ratio of PC:cholesterol:dicetyl phoshate) was investigated on silicon oxide interfaces [89].
PLA2 interaction with phospholipid bilayers (DPPC, DOPC, or POPC coadsorbed with dodecyl maltoside) at silica surfaces [90] has been studied [91]. The conventional model of PLA2–membrane interaction assumes adsorption of the enzyme on the bilayer, followed by partial extraction of a substrate molecule into the enzyme, hydrolysis and release of the products [92]. In distinction, neutron reflectometry results suggest that the enzyme actually penetrates into the layer until the active site, located at the top of the enzyme, is at the same level with the lipid headgroups of the outer phospholipid layer. The penetration of the enzyme into the lipid layer would then imply that the rate determining step of the overall reaction is either the initial adsorption or the final desorption, rather than the actual hydrolysis.
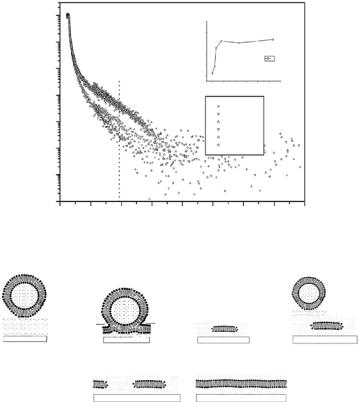
In an attempt to better control the formation of lipid bilayers on solid support, a sequential deposition of phospholipid monolayers in a combination of Langmuir–Blodgett and Langmuir–Schaefer transfer techniques has been utilized [93]. Neutron reflection measurements allowed precise, nondestructive characterization of the structure, hydration and roughness of the deposited layers. Beyond the first bilayer, deposited by Langmuir–Blodgett transfer, a second, “free” and fully hydrated bilayer was formed and its physical
14 Biomimetic Structures at Surfaces and Interfaces |
299 |
Reflectivity
1
0.1
0.01
1E-3
1E-4
1E-5
1E-6
1E-7
0.00
DMPC vesicle adsorption at Si− D2O interface
5.0 10−1
4.0 10−1
3.0 10−1
2.0 10−1
Reflectivity |
|
|
|
|
|
B |
|
1.0 10−1 |
|
|
|
|
|
|
|
|
0.0 |
|
|
|
|
|
|
|
0 |
100 |
200 |
300 |
400 |
500 |
600 |
|
|
|
Time (min) |
|
|
||
Si/D2O 0−20 min 20−32 min
68−80 min
170−230 min
470−530 min
0.05 |
0.10 |
0.15 |
0.20 |
0.25 |
0.30 |
0.35 |
0.40 |
|
|
q [Ang−1] |
|
|
|
||
D2O |
|
|
D2O |
|
|
|
|
|
D2O |
|
|
|
|
D2O |
D2O |
|
|
|
|
Quartz substrate |
Quartz substrate |
D2O |
D2O |
Quartz substrate |
Quartz substrate |
||
|
D2O |
D2O |
|
|
D2O |
D2O |
|
|
Quartz substrate |
Quartz substrate |
|
Fig. 14.8. Top: Neutron reflectivity demonstrating the adsorption of DMPC vesicles at an Si–SiO2 surface in D2O. The inset shows the change in reflected intensity at a fixed momentum transfer, Qz = 0.09 ˚A−1. Bottom: Schematic depiction of the adsorption of uni-lamellar phospholipid vesicles onto a solid hydrophilic interface [85]
properties examined (Fig. 14.9) [94]. Utilizing this model system, the insertion of a model peptide that was determined to be an active transmembrane shuttle for drug delivery to cells demonstrated uniform peptide distribution in the interfacial lipid region [95]. In the presence of dipalmitoyl phosphatidylserine (DPPS), the peptide was detected mainly in the lipid headgroup region and increased bilayer roughness was observed in the neutron reflectometry measurements.
14.6.2 Hybrid Bilayer Membranes
A variation of the theme of mimicking biomembranes on solid interfaces are the so-called hybrid bilayer membranes (HBMs). A lipid monolayer is fused onto a chemically grafted SAM or polymer layer [16, 96–98]. The basic details of this approach are outlined in the contribution by Majkrzak et al., in this volume.
300 T. Gutberlet et al.
(a)
(1) |
(2) |
(b)
Reflectivity
|
|
|
|
|
|
|
|
|
|
|
|
Dw |
|
|
(3) |
|
|
|
|
|
|
|
|
|
(4) |
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
10−1 |
|
|
|
|
|
|
|
|
|
|
24 |
|
|
|
|
|
|
|
|
|
|
|
|
22 |
|
|
|
10−2 |
|
|
|
|
|
|
|
|
|
(Å) |
20 |
|
|
|
|
|
|
|
|
|
|
|
Dw |
18 |
|
|
|
10−3 |
|
|
|
|
|
|
|
|
|
16 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
14 |
|
|
|
10−4 |
|
|
|
|
|
|
|
|
|
|
12 |
50 |
55 T (*C) |
|
|
|
|
|
|
|
|
|
|
45 |
|||
10−5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
10−6 |
(Å) |
6 |
|
|
|
|
|
|
|
|
|
|
|
4 |
|
|
|
|
|
|
|
|
|
|
|
||
10−7 |
thickness |
5 |
|
|
|
|
|
|
|
|
|
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
||
|
|
3 |
|
|
|
Dw |
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
|
|
|
|
10−8 |
|
0 |
20 |
40 |
60 |
80 |
100 |
120 |
140 |
|
|
|
|
|
0 |
|
|
|
|
||||||||
|
|
|
Thickness (Å) |
|
|
0.1 |
0.15 |
0.2 |
0.25 |
||||
0 |
|
|
0.05 |
|
|
||||||||
q (Å−1)
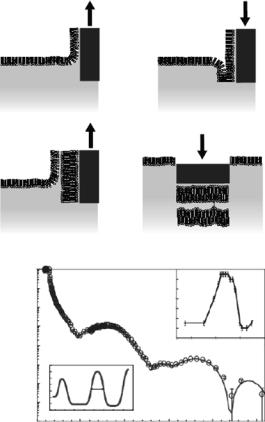
Fig. 14.9. (a) Cartoon demonstrating the four-step preparation of a double bilayer. Layers 1–3 are deposited by the Langmuir–Blodgett technique; layer 4 is deposited by the Langmuir–Schaefer method [93]. (b) Neutron reflectivity profile and fitted SLD profile (bottom left inset ) for a DSPC double bilayer in D2O at 59◦C. Upper inset: variation of the inter-bilayer distance (Dw) with temperature [94]
By preparing a HBM on a gold-coated Si wafer with an ultrathin (only several micrometer) aqueous subphase in a solid–liquid sample cell – to reduce incoherent background – neutron reflectivity data with a momentum transfer of 0.7 ˚A−1, spanning a dynamic range of 108, have been recorded [17]. This work showed that the peptide melittin perturbs the phospholipid headgroup strongly and also a ects the acyl chain region of the distal bilayer leaflet of the HBM [17]. Also the combination of an HBM and a subsequently transferred bilayer leads to the formation of a “free” phospholipid bilayer floating atop the HBM. Such systems have been studied with neutron reflectometry,
14 Biomimetic Structures at Surfaces and Interfaces |
301 |
and interpreted in the VRDF approach [97]. The free bilayer is organized according to the steric demands of the lipids rather than the influence of the substrate. The advantage of the free supported bilayer compared to more conventional supported systems is that the mobility of the phospholipid molecules makes them suitable for investigations of transmembrane processes. This was demonstrated by the incorporation of a deuterated peptide into the bilayer that a orded the observation of peptide orientational changes induced by small potential changes across the membrane structure [99].
Phospholipid monolayer formation on a hydrophobic polymer film (polystyrene, PS, spin-coated on a Si wafer) proceeds very fast after exposure of the surface to a vesicle suspension [98]. Contrast variation via D2O–H2O exchange and the use of deuterated compounds (DMPC-d54, deuterated PS) made the approach highly sensitive to the adsorbed layer structure, even minor contaminations or structural changes caused by membrane active molecules– such as proteins, DNA, or detergents–are readily accessible. Using this approach, the interaction of the neurotoxic β-amyloid peptide Aβ (25–35) with a phospholipid monolayer obtained via fusion of small unilamellar vesicles onto a PS-coated substrate has been studied. It was shown that the peptide adsorbed rather than penetrated the supported lipid monolayer [100], in contrast to results of neutron di raction experiments on phospholipid multilayer stacks, which indicated a deep penetration of Aβ amyloid into the lipid matrix [101].
14.6.3 Polymer-Supported Phospholipid Bilayers
Another problem in the context of membrane protein incorporation into biomembrane mimics is the nonphysiological interaction of the proteins with the solid support that would result in lateral immobilization or denaturation. The development of free bilayers or of bilayers that are tethered to the solid interface by means of polymer cushions has been proposed to solve this problem [1,102]. Promising systems that have been investigated for the preparation of polymer cushions have involved PEGs and polyethylenimines (PEIs). Also, polyelectrolyte (PE) multilayer films, where well-defined systems up to micrometer thickness may be obtained via sequential, layer-by-layer deposition of alternately charged PEs [104], have been employed. Their internal structure have been analyzed using neutron and X-ray reflectivity [48, 106].
The formation of DMPC bilayers on PEI-coated quartz substrates using a variety of deposition techniques has thus been studied by neutron reflection [103]. Using the layer-by-layer preparation technique, the attachment of individual phospholipid bilayers on poly(styrene sulfonate), PSS, and poly(allylamine hydrochloride), PAH, has been demonstrated [107–109]. Subsequently, DMPG bilayers on a PSS/PAH cushion were utilized to study the interaction of β-amyloid peptide with membranes [107]. In this work, neutron reflectometry demonstrated the presence of a 10 ˚A amyloid layer adsorbed to the phospholipid bilayer. Similarly, the in situ adsorption of a dense layer
302 T. Gutberlet et al.
of the enzyme, phosphatidylinositol-3-kinase (PI3K), to phosphatidic acid on PAH/PSS, was followed by neutron reflection [109]. In another study, the adsorption of pectin onto charged and uncharged lipid bilayers on PE cushions was investigated [108]. Recently, neutron reflectometry was applied to study the interaction of DNA with PE-supported DMPC and DMPG bilayers in situ [110]. At high phospholipid concentrations (>0.5 mg ml−1), the adsorption of multilamellar stacks of DMPC on PSS/PAH cushions was observed [111]. A very recent study demonstrated the option to deposit further PE layers on top of a DMPC bilayer which in turn was adsorbed to PE multilayer cushions [112]. This approach might lead to PE encapsulated biomembrane mimics with potential application for novel drug delivery systems. A PE cushion terminated with a terpolymer that links stearoyl chains to a styrolsulfonate anchor was created by deposition of positively charged poly-l- lysine and negatively charged alginate. Subsequent transfer of a phospholipid monolayer resulted in the construction of a tethered hybrid bilayer membrane whose structure was in detail assessed by neutron reflectometry [113].
14.7 Conclusions
Neutron reflectometry provides valuable insights into the structure and assembly of complex biological systems at fluid surfaces and solid–liquid interfaces. The examples described in this chapter were selected to demonstrate current capabilities. For a more detailed characterization of biomimetic systems, on the other hand, not only organization perpendicular to the membrane surface is required, but also lateral structures, membrane in-plane organization and dynamic membrane features [1, 114]. Improvements in grazing-incidence neutron di raction and o -specular neutron reflectivity are requested to tackle these questions. Investigations by these techniques on phospholipid membranes as outlined in the contribution by Salditt et al., in this volume and pioneering studies by Huang and coworkers [115,116] demonstrate the feasibility of these approaches, which currently, however, still lack su cient neutron flux.
Acknowledgments
M.L. wishes to acknowledge support by the Volkswagen Foundation (Grant no. I/77709), the National Institutes of Health (Grant no. 1 RO1 RR14812) and The Regents of the University of California.
References
1.E. Sackmann, M. Tanaka, Trends Biotechnol. 18 (2000), 58–64
2.M. L¨osche, in Current Topics in Membranes, Vol. 52, S.A. Simon, T.J. McIntosh (Eds.) (Academic Press, New York, 2001)
3.G. Fragneto-Cusani, J. Phys.: Condens. Matter 13 (2001), 4973–4989
14 Biomimetic Structures at Surfaces and Interfaces |
303 |
4.C.F. Majkrzak, N.F. Berk, Appl. Phys. A 74 (2002), S67–S69
5.R. Cubitt, G. Fragneto, R.E. Gosh, A.R. Rennie, Langmuir 19 (2003), 7685– 7867
6.R. Frahm, J. Weigelt, G. Meyer, G. Materliki, Rev. Sci. Instrum. 66 (1995), 1677–1680
7.J.F. Ankner, C. Rehm, Physica B 336 (2003) 68–74
8.K. Lieutenant, H. Fritzsche, F. Mezei, Appl. Phys. A 74 (2002), S1613–S1615
9.P. Kr¨uger, M. L¨osche, in Lecture Notes in Physics, Vol. 634, R. Haberlandt,
D.Michel, A. P¨oppl, R. Stannarius (Eds.) (Springer, Berlin–New York, 2004)
10.J. Als-Nielsen, D. Jacquemain, K. Kjaer, M. Lahav, F. Leveiller, L. Leiserowitz, Phys. Rep. 246 (1995), 251–313
11.M. L¨osche, J.P. Rabe, A. Fischer, B.U. Rucha, W. Knoll, H. M¨ohwald, Thin Solid Films 117 (1984), 269–280
12.M. L¨osche, E. Sackmann, H. M¨ohwald, Ber. Bunsenges. Phys. Chem. 87 (1983) 848–852
13.P. Kr¨uger, M. L¨osche, Phys. Rev. E 62 (2000), 7031–7043
14.M. L¨osche, H. M¨ohwald, Rev. Sci. Instrum. 55 (1984), 1968–1972
15.P.S. Pershan, Colloids Surf. A: Physicochem. Eng. Aspects 171 (2000), 149– 157
16.C.W. Meuse, S. Krueger, C.F. Majkrzak, J.A. Dura, J. Fu, J.T. Connor, A.L. Plant, Biophys. J. 74 (1998), 1388–1398
17.S. Krueger, C.W. Meuse, C.F. Majkrzak, J.A. Dura, N.F. Berk, M. Tarek, A.L. Plant, Langmuir 17 (2001), 511–521
18.M.C. Wiener, S.H. White, Biophys. J. 59 (1991), 174–185
19.D. Vaknin, K. Kjaer, J. Als-Nielsen, M. L¨osche, Biophys. J. 59 (1991), 1325– 1332.
20.M. Schalke, P. Kr¨uger, M. Weygand, M. L¨osche, Biochim. Biophys. Acta 1464 (2000), 113–126
21.M. Schalke, M. L¨osche, Adv. Colloid Interf. Sci. 88 (2000), 243–274
22.R.S. Armen, O.D. Uitto, S.E. Feller, Biophys. J. 75 (1998), 734–744
23.H.L. Brockman, Curr. Opin. Struct. Biol. 9 (1999), 438–443
24.J. Als-Nielsen, H. M¨ohwald, in Handbook on Synchrotron Radiation, Vol. 4, S. Ebashi, M. Koch, E. Rubenstein (Eds.) (Elsevier, North Holland, 1991) pp. 1–53
25.G.S. Smith, J. Majewski, in Lipid Bilayers, Structure and Interactions, J. Katsaras, T. Gutberlet (Eds.) (Springer, Berlin, 2001) pp. 127–148
26.G. B¨uldt, H.U. Gally, A. Seelig, J. Seelig, G. Zacchai, Nature 27 (1978), 182– 184
27.G. B¨uldt, H.U. Gally, J. Seelig, G. Zacchai, J. Mol. Biol. 134 (1979), 673–691
28.T.M. Bayerl, R.K. Thomas, J. Penfold, A.R. Rennie, E. Sackmann, Biophys.
J.57 (1990), 1095–1098.
29.T. Brumm, C. Naumann, E. Sackmann, A.R. Rennie, R.K. Thomas, D. Kanellas, J. Penfold, T.M. Bayerl, Eur. Biophys. J. 23 (1994), 289–295.
30.C. Naumann, C. Dietrich, J.R. Lu, R.K. Thomas, A.R. Rennie, J. Penfold, T.M. Bayerl, Langmuir 10 (1994) 1919–1925.
31.C. Naumann, T. Brumm, A.R. Rennie, J. Penfold, T.M. Bayerl, Langmuir 11 (1995) 3948–3952.
32.S.J. Johnson, T.M. Bayerl, W. Weihan, H. Noack, J. Penfold, R.K. Thomas,
D.Kanellas, A.R. Rennie, E. Sackmann, Biophys. J. 60 (1991), 1017–1025
33.B. Dem´e, D. Hess, M. Tristl, L.T. Lee, E. Sackmann, Eur. Phys. J. E 2 (2000), 125–136
304T. Gutberlet et al.
34.C. Naumann, C. Dietrich, A. Behrisch, T.M. Bayerl, M. Schleicher, D. Bucknall, E. Sackmann, Biophys. J. 71 (1996), 811–823
35.J. Strzalka, B.R. Gibney, X. Chen, C.C. Moser, P.L. Dutton, S.K. Satija, B.M. Ocko, J.K. Blasie, Biophys. J. 78 (2000), 325A
36.M. Kent, H. Yim, D. Sasaki, J. Majewski, G.S. Smith, K. Shin, S. Satija, B.M. Ocko, Langmuir 18 (2002), 3754–3757
37.M. Kent, H. Yim, D. Sasaki, S. Satija, J. Majewski, T. Gog, Langmuir 20 (2004), 2819–2829
38.W.K. Fullagar, K.A. Aberdeen, D.G. Bucknall, P.A. Kroon, I.R. Gentle, Biophys. J. 85 (2003), 2624–2632
39.W.K. Fullagar, I.R. Gentle, S.A. Holt, ISIS2003 Science Highlights
40.U.B. Sleytr, M. S´ara, D. Pum, B. Schuster, Progr. Surf. Sci. 68 (2001), 231–278
41.B. Wetzer, A. Pfandler, E. Gy¨orvary, D. Pum, M. L¨osche, U.B. Sleytr, Langmuir 14 (1998), 6899–6706
42.A. Diederich, C. Sponer, D. Pum, U.B. Sleytr, M. L¨osche, Colloids Surf.
B:Biointerf. 6 (1996), 335–346
43.M. Weygand, B. Wetzer, D. Pum, U.B. Sleytr, N. Cuvillier, K. Kjaer, P.B. Howes, M. L¨osche, Biophys. J. 76 (1999), 458–468
44.M. Weygand, M. Schalke, P.B. Howes, K. Kjaer, J. Friedmann, B. Wetzer, D. Pum, U.B. Sleytr, M. L¨osche J. Mater. Chem. 10 (2000), 141–148
45.M. Weygand, K. Kjaer, P.B. Howes, B. Wetzer, D. Pum, U.B. Sleytr, M. L¨osche, J. Phys. Chem. B 106 (2002), 5793–5799
46.D. Vaknin, J. Als-Nielsen, M. Piepenstock, M. L¨osche, Biophys. J. 60 (1991),
1545–1552
47. D. Vaknin, K. Kjaer, H. Ringsdorf, R. Blankenburg, M. Piepenstock,
A. Diederich, M. L¨osche, Langmuir 9 (1993), 1171–1174
48.M. L¨osche, M. Piepenstock, A. Diedrich, T. Grunewald, K. Kjaer, D. Vaknin, Biophys. J. 65 (1993), 2160–2177
49.M. L¨osche, C. Erdelen, E. Rump, H. Ringsdorf, K. Kjaer, D. Vaknin, Thin Solid Films 242 (1994), 112-117
50.M. Tristl, I. Haase, S. Marx, L.T. Lee, M. Fischer, E. Sackmann, LLB Sci. Rep. (1999-2000) 100–102; K. Sengupta, L. Limozin, M. Tristl, M. Fischer, E. Sackmann, Phys. Chem. Chem. Phys., submitted
51.C.E. Miller, J. Majewski, R. Faller, S. Satija, T.L. Kuhl, Biophys. J. 86 (2004), 3700–3708
52.T.L. Kuhl, J. Majewski, J.Y. Wong, S. Steinberg, D.E. Leckband, J.N. Israelachvili, G.S. Smith, Biophys. J. 75 (1998), 2352–2362
53.J. Majewski, T.L. Kuhl, M.C. Gerstenberg, J.N. Israelachvili, G.S. Smith, J. Phys. Chem. B 101 (1997), 3122–3129
54.A. Wurlitzer, E. Politsch, S. H¨ubner, P. Kr¨uger, M. Weygand, K. Kjaer, P. Hommes, O. Nuyken, G. Cevc, M. L¨osche, Macromolecules 34 (2001), 1334– 1342
55.T. Gutberlet, A. Wurlitzer, U. Dietrich, E. Politsch, G. Cevc, R. Steitz, M. L¨osche, Physica B 283 (2000), 37–39
56.E. Politsch, G. Cevc, A. Wurlitzer, M. L¨osche, Macromolecules 34 (2001), 1328–1334
57.E. Politsch, J. Appl. Crystallogr. 34 (2001), 239–251
58.C. Hiergeist, R. Lipowsky, J. Phys. II France 6 (1996), 1465–1481
59.A. Baszkin, W. Norde (Eds.), Physical Chemistry of Biological Interfaces
(Dekker, New York, 1999)
