
Neutron Scattering in Biology - Fitter Gutberlet and Katsaras
.pdf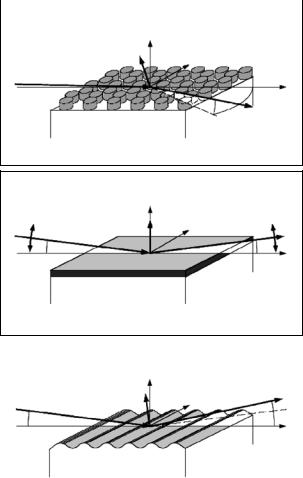
14 |
Biomimetic Structures at Surfaces and Interfaces |
285 |
||
|
|
|
|
|
(a) |
|
diffraction at |
|
|
grazing incidence (GID)
z
|
|
Q |
|
|
|
|
|
x |
|
|
a0 |
|
|
|
k0 |
|
|
||
|
|
|
y |
|
|
|
|
ar |
2Θ |
|
|
|
lxy,z |
|
|
|
|
|
kxy,z |
|
a0 100 mdeg |
2D crystalline order |
||
|
|
|
||
|
I(Q ) = IBragg (Q ) |
¤ within interface layer |
||
(b)
|
|
|
|
z |
|
|
|
|
Qz = 2|k0| sin α |
||
k0 |
||
l0 |
a0 |
|
|
||
|
ar = a0 |
|
|
R = lr / l0 = R(Qz ) |
specular reflection
|
|
|
kr |
||
x |
||
ar |
lr |
|
|
||
|
y |
¤film structure normal to interface
(c) |
|
near-specular diffuse scattering |
||
|
|
|
|
|
|
|
z |
|
|
|
|
Q |
|
|
|
|
kr |
lr |
|
|
|
|||
|
k0 |
x |
|
|
|
l0 |
|
ar |
|
|
a0 |
|
|
|
|
|
|
|
y |
|
ar ~ a0 |
¤ correlated defects |
|
|
|
lr = lr (Q ) |
|
||
|
within interface |
|
|
|
Fig. 14.1. Surface-sensitive scattering experiments at grazing incident angles [2]
the “virtual” resolution. One of the most striking advantages, reflectivity measurements do not require crystalline samples to obtain high resolution, and are thus capable of probing membraneous systems in their native, disordered state.
To achieve the highest possible resolution of a structure in reflectivity measurements, there are strict requirements on the sample with respect to the quality of the interface and to in-plane sample homogeneity. Clearly, to achieve subnanometer resolution on a sample film, the substrate has to be of ideal geometry down to the nanometer scale: interfaces with residual rms
286 T. Gutberlet et al.
roughnesses in the ˚Angstrom regime are generally required. Moreover, a microscopically smooth but macroscopically curved sample surface also reduces resolution. This is one of the reasons why reflectivity measurements from systems at fluid surfaces have been relatively successful. Due to the action of gravity, a free water surface is “by definition” perfectly flat on the macroscopic length scale. On the microscopic length scale, thermally excited capillary waves, controlled by a competition between surface tension and thermal excitation, define the quality of the fluid substrate. For water with its high surface tension, the residual roughness is on the order of 3 ˚A – perfectly suited for high-quality reflection measurements. A second reason fluid surface monolayers have been frequently studied with neutron scattering is that the free fluid surface permits easy access and control of most relevant systems parameters, such as molecular density within the film and subphase chemistry and temperature, etc. Moreover, in situ manipulation, e.g., the injection of peptides or proteins underneath a previously prepared and characterized lipid film, is straight-forward as shown further below.
Another concern is in-plane homogeneity of the samples. Solid-substrate borne monolayer systems – so-called “Langmuir–Blodgett” films [11] – as well as lipid surface monolayers, particularly within a first-order phase transition [12], show frequently domain structures on the micrometer scale [13]. Since phase information is lost in the determination of the scattered intensity, the experimentally measured reflectivity cannot be directly inverted to obtain an SLD profile. The usual work-around is data modelling, i.e., to parameterize the SLD and optimize parameters by fitting to the experimental data. For this process to be tractable, in-plane heterogeneities have to be avoided, as they increase largely the number of model parameters that describe the microstructure. It is thus important that the sample preparation protocol ensures that films are homogeneous on the length scale of the in-plane coherence length of the probe beam. In angle-dispersive reflectivity measurements, this coherence length is determined by the geometry of the instrument and is typically largely anisotropic within the film plane – on the order of 105 ˚A in the projected direction of the beam onto the sample and on the order of 100 ˚A perpendicular to this direction. If one takes a geometric mean of these numbers as a criterion, then the sample needs to be homogeneous on the micrometer length scale. This quantification suggests that optical microscopy [14] is an appropriate technique to check routinely the suitability of samples for reflectivity characterization.
Given in-plane homogeneity of the sample and optimal substrate geometry, the ultimate limitation to resolution derives from the steep drop of the scattered intensity as a function of momentum transfer, Qz (see e.g., Majkrzak et al., in this volume). The reflected intensity R drops as Q4z for Qz Qcz , the critical momentum transfer of total reflection. This results in R being vanishingly small, on the order of 10−6–10−8 once Qz ≥ 0.3 ˚A−1 for X-ray or neutron measurements of aqueous surface films. Clearly, X-ray probes, particularly at high-brilliance third-generation synchrotron sources, have great advantage
14 Biomimetic Structures at Surfaces and Interfaces |
287 |
over neutrons as it comes to determining this one scattered probe particle out of 106 incident probes! Ultimately, however, the limits of resolution cannot be indefinitely pushed to ever higher Qz values by using ever-increasing primary beam intensities. Not only is beam damage a serious issue in X-ray scattering at synchrotron sources, but also both near-specular scattering from capillary waves [15] and incoherent scattering from the bulk phase – excited by the impinging beam which penetrates deeply into the bulk at high Qz , and hence high incident angles – limit the detection of the minute number of specularly reflected probes in practical terms.
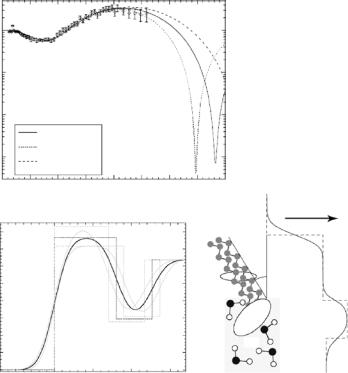
Typical maximum Qz values amount to 0.8 ˚A−1 in X-ray reflection and 0.3–0.6 ˚A−1 in neutron reflection measurements, largely depending on the isotopic nature of aqueous bulk phase and design of the sample cell. To reduce incoherent background subphase thickness needs to be kept at an absolute minimum [16, 17]. As the sample theorem would suggest, such Qmaxz values correspond to a “canonical” [18] resolution of ∆z = π/Qmaxz ≈ 5–10 ˚A. The practical consequences are illustrated in Fig. 14.2. Based on a single neutron reflectivity measurement that spans the range out to Qz ≈ 0.25 ˚A−1, it is not even possible to determine the thickness of the hydrophobic slab within a lipid surface monolayer to any reasonable certainty – even as the methylenes are fully deuterated.
14.2.2 Structure-Based Model Refinement
Fortunately, this situation has been remedied by the development of structurebased model refinement techniques [18–21]. “Composition-space refinement” [18] takes the parameterization from a level where one describes 1D SLD profiles to a level where one parameterizes the molecular structure of the interface architecture [18, 19]. This can be implemented in terms of the atomic content of the slabs in a box model [19] or, more directly, in terms of thermally broadened distributions of molecular subfragments. One realization of the latter approach is a “volume-restricted distribution function” (VRDF) parameterization [20, 21], in which the molecular fragments are subject to the condition that they just fill the available space. Thus, volumetric information on the molecular subfragments is essential and is usually derived from molecular dynamics simulations [22]. Since this approach determines the packing of molecular subfragments within the plane of the monolayer, the retrieved information is “quasi-2D” in character.
There are various benefits of the VRDF approach to modelling reflectivity data:
–At a resolution of π/Qmaxz better than ≈5 ˚A, the description of a monolayer even of the simplest phospholipid in terms of a homogeneous headgroup slab breaks down within the conventional box model: significant discrepancies between models and data that are frequently encountered at high Qz [21]. VRDF models reconcile these discrepancies.
288 T. Gutberlet et al.
(a)
F |
|
|
R/R |
1 |
|
reflectivity, |
||
10−1 |
||
|
||
neutron |
10−2 |
|
nomalized |
||
10−3 |
||
|
DPPC−d62 on D2O, π =42 mN/m
dchain = 16.0 Å
dchain = 18.0 Å
dchain = 14.0 Å
(b) |
) |
|
2− |
|
Å |
|
6− |
|
(10 |
|
n |
|
rneutronSLD, |
0 |
0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.35 |
0.4 |
momentum transfer, Qz (Å−1)
electron density
8 |
|
|
|
|
7 |
|
|
|
|
6 |
|
|
|
Alipid |
|
|
|
|
|
5 |
|
|
|
b |
4 |
|
|
|
phos- |
3 |
|
|
|
|
|
|
|
phatidly- |
|
2 |
|
|
|
choline |
|
|
|
H2O |
|
1 |
|
|
|
|
|
|
|
|
|
0 |
|
−10 |
−20 |
−30 |
10 |
0 |
distance to interface, z (Å)
Fig. 14.2. Qualitative assessment of the information content of a reflection spectrum. Shown is a typical neutron reflectivity data set, R/RF (chain-perdeuterated DPPC-d62 on D2O at π = 42 mN m−1 [19]) (a) in comparison with various model reflectivities (b). A scheme of the molecular arrangement at the interface is shown to the right. In a combination of X-ray and neutron reflection measurements, the hydrophobic layer thickness has been determined to be dhphob = 16.0 ˚A. The corresponding scattering length density (SLD) profile is shown as a bold line. To assess the information content of the single neutron reflection data set shown, detuning of dhphob by ±2 ˚A and readjusting the corresponding SLD and surface roughness to fit the DPPC-d62/D2O data leads to models – shown as dashed lines – that cannot be discriminated within the resolution of the single experiment. However, a combination of X-ray and neutron scattering is very well capable of resolving the structure, even as the resolution of every single experiment is insu cient. Adapted from [21]
14 Biomimetic Structures at Surfaces and Interfaces |
289 |
–The parameterization of the system chemistry permits a strict coupling of di erent data sets – obtained for various isotopic contrasts or neutron and X-ray measurements on isomorphic systems – thus boosting the e ective resolution greatly.
–Finally, even if only one data set for a single contrast is available (e.g., one single high-resolution X-ray reflectivity data set), the chemically intuitive parameterization permits an intuitive molecular-level interpretation of the results – an advantage over multilayer box models which do not generally lend themselves to a straight-forward interpretation in terms of a chemical structure.
In Section 14.4, below results on various classes of supramolecular systems at aqueous surfaces and interfaces are collected to enable the reader to judge what information may be retrieved with surface-sensitive neutron scattering in various areas of bioinspired materials science.
14.3 Floating Lipid Monolayers:
Structural Investigations and the Interaction of Peptides and Proteins with Lipid Interfaces
Amphiphilic molecules, such as phospholipids, self-assemble into free-floating monolayers (“Langmuir monolayers”, LM) at the air–water interface due to their hydrophilic headgroup linked to hydrophobic acyl chains. As mentioned above, LMs are physicochemically well-controlled model systems, which provide unique opportunities for the investigation of structure and interaction of a biomembrane mimic with biomolecules, e.g., peptides and proteins [23]. Consequently, LMs have been early on studied in X-ray and neutron scattering experiments [24, 25] that have revealed their SLD distribution along the surface normal, indicative of their time-averaged thermally broadened structure.
The relatively low resolution in neutron reflectivity experiments is compensated for by isomorphic contrast variation via exchange of protons with deuterium in biomolecules, which exploits the large di erence in neutron scattering length of the two isotopes (see also contribution by Lu, in this volume). Molecular subunits within a monolayer, embedded or adsorbed molecules – such as surfactants, peptides or proteins – or phase separation within the layer plane can be characterized by choosing appropriate contrasts in multiple scattering experiments. In pioneering work using neutron di raction, this was utilized for the localization of methylene segments in the hydrophobic region and headgroup orientation of selectively deuterated phospholipid bilayers [26, 27].
290 T. Gutberlet et al.
14.3.1 Single Phospholipid LMs
The structure and organization of single phospholipid monolayers on aqueous subphase using di erent SLD contrasts have been investigated in several studies with neutron reflectometry [19,28–31]. The most convenient way to achieve contrast variation in LM systems is by preparation of the monolayer aqueous subphase with either H2O or D2O. The adsorption of protonated molecular species at the monolayer–D2O interface decreases the SLD there substantially and is sensitively detected in a reflectivity experiment. Thus, the electrostatic coupling of poly-l-lysine to DMPC monolayers and to DMPC–dimyristoyl phosphatidylglycerol (DMPG) mixed monolayers was studied on D2O subphase. Similarly, the penetration of the protein spectrin into the monolayer headgroup region was characterized [32] and the adsorption of F-actin filaments to cationic LMs of dimyristoyl trimethylammonium propane (DMTAP) was investigated [33].
Another convenient method to vary the contrast in LM systems is by using lipids with perdeuterated hydrophobic chains. The interaction of hisactophilin with the lipid interface was explored [34]. The protein, which has a myristic acyl chain anchor attached, binds tightly to negatively charged membranes. Changes in the SLD profile upon penetration of the protonated myristic acid anchor into the deuterated lipid matrix, distinguished adsorption of protein from penetration of protein into the lipid interface, thus discriminating electrostatically and hydrophobically driven lipid–protein interaction. With acyldeficient hisactophilin only the formation of a protein layer adsorbed was observed beneath the charged phospholipid monolayer that retained its SLD at the value prior to protein exposure. For a fully synthetic system, the orientation of a palmitoylated diα-helical peptide with di erent deuterium-labeled positions beneath a LM of dilaurylphosphatidylethanolamine (DLPE) was examined [35] in combined X-ray and neutron reflectivity experiments. Similarly, the interaction of myoglobin with lipid monolayers was investigated with a combination of neutron and X-ray reflection and GIXD [36, 37]. Underneath mixed monolayers of DPPC/PSIDA, a diacylglycerol lipid with a metal-chelating imino diacetate headgroup, the formation of a densely packed monolayer of myoglobin was observed with thickness and density consistent with crystallographic data for myoglobin. With Cu2+ ions in the subphase, the protein adsorbed in a side-on orientation at low packing density but tilted to an end-on orientation at high packing density. With Ni2+ ions, on the other hand, myoglobin was observed to adsorb in an end-on orientation at all densities. These di erences were attributed to a stronger interaction of the histidine moieties with the imino diacetate anchor mediated by Cu2+.
A number of studies have been conducted to understand the structure and function of components lining the interface of the alveoli, known as lung surfactant [38]. The lipid layer consists mainly of a mixture of DPPC and the anionic lipid dipalmitoylphosphatidylglycerol (DPPG) and forms a matrix for four surfactant-associated proteins (SP-A, SP-B, SP-C, and SP-D).
14 Biomimetic Structures at Surfaces and Interfaces |
291 |
X-ray reflectivity measurements on surfactant-associated protein monolayers have been performed in conjunction with pressure–area isotherms and modelling to suggest that the protein undergoes changes in its tertiary structure at the air–water interface under the influence of surface pressure variations, indicating the likely role of such changes in surfactant squeeze out as well as lipid exchange between the air–alveoli interface and the underlying subphase. Recent neutron reflectivity data on bovine SP-B monolayers on aqueous subphases are consistent with the exchange of a large number of labile protons as well as the inclusion of a significant amount of water, which is partly squeezed out of the protein monolayer at elevated surface pressures [38]. In a mixed SP-B and DPPC system, squeeze out of the protein and re-adsorption upon compression and re-expansion was observed, with most of the protein material predominantly associated with the interface. Only small quantities of lipid followed the protein on leaving the monolayer at compression as shown by comparison of SLD profiles with protonated and deuterated DPPC [39].
Bacterial S-layer proteins [40] crystallize at a wide range of interfaces and surfaces, including phospholipid membranes [41]. The microscopic interactions between recrystallized bacterial S-layers and floating phosphatidylethanolamine (PE) LMs have been analyzed using FT-IRRAS spectroscopy, X-ray reflectivity and grazing-incidence di raction, and neutron reflectivity at air-water interfaces [42–44]. A slight increase of the lipid acyl chain order was observed in GIXD upon protein adsorption, indicative of an increase in local lipid density. Corefinement of X-ray and neutron reflectivity data suggested that protein interpenetrates the lipid monolayer only in the headgroup region. Since only a small amount of protein material is observed within the headgroups, it was inferred that structural intact protein motifs enter the PE headgroups at localized, and possibly repetitive, interaction points within the S-layer crystal lattice – rather than a laterally homogeneous interaction of the protein occurring with the lipid surface monolayer [45].
14.3.2 Functionalized Phospholipid LMs
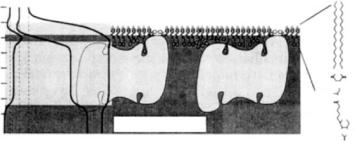
Various studies utilized the strong specific binding of biotin to streptavidin to probe protein interactions with functionalized lipid monolayers on aqueous subphases [46–49]. Fluorescence microscopy using FITC-labeled streptavidin showed that the protein forms – presumably crystalline – domains and established preparation conditions where these domains covered the surface quantitatively. Neutron reflection experiments showed the formation of a monomolecular protein layer with an e ective thickness of 44 ± 2 ˚A. Quantitative binding was observed already at ultralow biotin surface concentrations. A combination of X-ray and neutron scattering experiments subsequently revealed distortions of the LM by the tightly bound protein (Fig. 14.3) [47]. Introducing a spacer between the functionalized lipid surface anchors and the biotin moiety resulted in a hydration layer between streptavidin and the lipid monolayer [49], concomitant with a reduction of monolayer distortion.
292 T. Gutberlet et al.
10
0 |
|
|
|
air |
|
|
|
|
|
||
−10 |
|
|
|
|
|
−20 |
|
|
|
|
|
−30 |
|
|
|
|
|
−40 |
|
|
|
|
|
−50 |
|
|
|
|
|
−60 |
n |
n |
x |
Water subphase |
|
Å |
H |
D |
H |
||
|
O O
O O
S
HOOC N
O
S
N N
O
Fig. 14.3. Schematic representation of streptavidin interaction with a biotinylated monolayer. The lipid monolayer is distorted by the tightly bound protein. SLD profiles are shown on the left (n, neutrons; x, X-rays; H, H2O; D, D2O) [47]
The binding dynamics of the recombinant protein, lumazine synthase at biotinylated lipid monolayers were studied with neutron reflectometry [50]. The protein was biotinylated and coupled to streptavidin. The binding of these constructs to biotinylated LMs was monitored by following the neutron reflection. A model including a densely adsorbed protein monolayer and progressively more dilute protein layers was used to describe the experimental data.
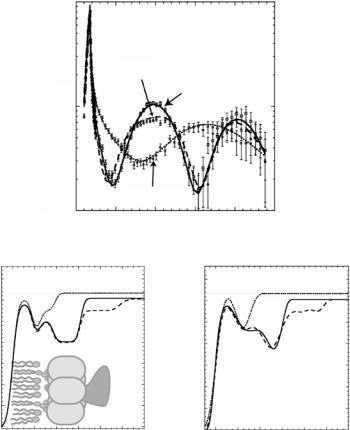
Recently neutron reflectometry was applied to characterize the structure of ganglioside GM1-functionalized LMs upon binding of cholera toxin (CTαβ5) and its β subunit (CTβ5) [51]. Structural parameters such as the density and thickness of the lipid layer, extension of the GM1 headgroup, orientation and position of the protein upon binding were inferred from the data. As shown in Fig. 14.4, the α subunit of the toxin was observed to be located underneath the pentameric ring of the B subunit and the CTβ5 is not penetrating into the lipid layer.
14.4 Lipopolymers
The grafting of water-soluble polymers to phospholipid membranes o ers an opportunity for a targeted modification of the surface properties of, e.g., liposomes. Depending on the grafting density, the organization of polymer chains at interfaces may vary between a “mushroom”-like conformation and an extended, ‘brush’-like configuration of the molecules (“polymer brush”) [52]. Such polymer brushes might act as soft, flexible cushions between adsorbed biomolecules and solid interfaces in applications that depend on surfacemodification with biopolymers.
The most prominent and best studied examples of such polymer brushes at lipid interfaces are comprised of polyethylene glycols (PEGs) as water soluble polymers that are chemically grafted to a phospholipid headgroup. Neutron
(b)
] −2
lengthScattering density, [Å
14 Biomimetic Structures at Surfaces and Interfaces |
293 |
(a)10−7
Reflectivity data
|
4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
z |
|
|
|
CTAB5 |
|
|
|
|
|
|
|
|
|
|
Q |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* |
|
|
|
|
|
CTB5 |
|
|
|
|
|
|
|
|
Reflectivity |
10−8 |
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
GM1:d-DPPE |
|
|
|
|
|
|
|
||
|
|
10−9 |
|
0.5 |
0.1 |
0.15 |
|
0.2 |
0.25 |
|
|
|||
|
|
|
0 |
|
|
|
|
|||||||
|
|
|
|
|
|
Qz [Å−1] |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
(c) |
|
|
|
|
|
|
|
|
7 10−6 Box model fit |
|
|
|
] |
7 10−6 |
Model independent fit |
|
|
||||||
|
|
|
GM1:d-DPPE |
2− |
|
|
|
|
|
GM1:d-DPPE |
|
|||
6 10−6 |
|
|
[Å |
6 10−6 |
|
|
|
|
||||||
|
CTB5 |
|
|
|
|
CTB5 |
|
|
||||||
5 10−6 |
|
CTAB5 |
density, |
|
|
|
|
CTAB5 |
||||||
|
|
|
5 10−6 |
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|||||||
4 10−6 |
|
|
|
|
|
length |
4 10−6 |
|
|
|
|
|
|
|
3 10−6 |
|
|
|
|
|
3 10 |
−6 |
|
|
|
|
|
|
|
1 10−6 |
|
|
|
|
|
Scattering |
|
|
|
|
|
|
|
|
|
|
|
|
|
1 10−6 |
|
|
|
|
|
|
|||
2 10−6 |
|
|
B5 |
A |
|
|
2 10 |
−6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
0 |
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
0 |
20 |
40 |
60 |
80 |
100 120 |
|
0 |
20 |
40 |
60 |
80 |
100 120 |
||
|
|
|||||||||||||
|
|
Length, Z [Å] |
|
|
|
|
|
|
Length, Z [Å] |
|
||||
Fig. 14.4. Neutron reflectivity of GM1/d-DPPE monolayer, monolayer with bound CTβ5, and monolayer with bound CTαβ5 (a). Lines indicate the fits corresponding to the SLD profiles from box-model fits in (b). The α subunit resides below the β pentamer, facing away from the lipid layer. An alternate set of SLD profiles from a model-independent spline fitting routine is shown in (c). The corresponding fits to the data were slightly better than the box-model fits [51]
reflectometry was applied to study the organization and structure of such PEGylated phospholipid monolayers in detail at the air–water interface [53]. Mixed with distearoylphosphatidylethanolamine (DSPE), a PEGylated lipid with 45 EG units exhibited the mushroom-to-brush transition upon monolayer compression, i.e., increasing polymer grafting density. The structure of the monolayer at the air–water interface was greatly perturbed by the presence of the bulky PEG–lipid headgroup resulting in a large increase of the apparent
294 T. Gutberlet et al.
thickness of the headgroup region normal to the interface and a concomitant roughening of the interface.
Using X-ray and neutron reflectivity, monolayers of short chain poly(methyl oxazoline) (PMeOx) attached to a diacylglycerol lipid anchor have been studied as neat systems [54] and in binary mixtures with DMPC [55]. A new data evaluation method was developed in the course of that work to specifically evaluate the structural organization of linear polymers at surfaces and interfaces [56, 57]. In this approach, the molecular structure of the polymer is described in terms of its scattering length and specific volume per unit length, and its configuration is parameterized in terms of inclination angles between rigid rods that represent the polymer chain. While this is already overparameterizing the problem, a whole ensemble of lipopolymer molecules is encoded for the data evaluation in terms of a large set of angles and other structural parameters of interest, thus deliberately overparameterizing the model even more. Working with this model on a multitude of X-ray and neutron data, functionally relevant configurations may be identified by an evolution–strategy algorithm. In the case of PMeOx, a phase transition upon film compression has been attributed to a tensile stress exerted by the chains on their anchor points [58] that leads to a partial immersion of the lipid anchors into the aqueous subphase by a few ˚Angstrom. This in turn results in a local condensation of the hydrophobic chains.
14.5 Protein Adsorption and Stability
at Functionalized Solid Interfaces
Adsorption and organization of biomolecules at solid interfaces bears implications to a broad range of areas, such as tissue engineering, biocompatibilization, and biosensorics [1]. Cell adhesion, cell–cell interaction, unfolding and denaturation of proteins are other important objectives related to the interaction of biomolecules with interfaces (for a comprehensive review see [59]). A number of aspects of protein adsorption to liquid and solid interfaces investigated with neutron reflectometry are already presented in the contribution by Lu, in this volume. Here we will discuss only investigations on protein interactions with surface-modified interfaces relevant for the construction of biosensors and biocompatible interfaces.
14.5.1 Hydrophobic Modified Interfaces
A well-established modification of native silicon–silicon oxide interfaces is silanization via e.g., octadecyltrichlorosilane (OTS) to hydrophobize the surface [52]. The structure and composition of bovine β-casein was investigated upon adsorption to OTS-modified silica surfaces in aqueous bu er [60, 61].
