
Molecular and Cellular Signaling - Martin Beckerman
.pdf
17
Cell Regulation in Bacteria
Bacteria are not only highly efficient organisms, but they are also the unseen majority wherever they exist. Bacteria are found in large numbers in the open oceans (1.2 ¥ 1029), in soils (2.6 ¥ 1029), in oceanic subsurfaces (3.5 ¥ 1030), and in terrestrial subsurfaces (~1 ¥ 1030). Each human body hosts some 1013 bacteria. One of the locales of high concentration is the gut. From 500 to 1000 different species of bacteria populate the human intestinal tract. The greatest concentration of bacteria is in the colon where there are about 1011 bacteria per gram of bulk material. This number is far greater than the amount of bacteria living on the surface of the skin. From 1000 to 10,000 bacteria live on each square centimeter of skin. These numbers yield a total skin population of about 300,000,000 bacteria.
Most of the bacteria that reside in the gastrointestinal (GI) tract are anaerobic, harmless, and even beneficial. One of the species of resident bacteria is the Gram-negative bacterium Bacteroides thetaiontaomicron. This bacterium acquires and hydrolyzes dietary polysaccharides that would otherwise be indigestible. Bacteroides thetaiontaomicron possesses a large number of genes that encode signaling proteins and gene regulators allowing the bacteria to communicate with and regulate their local environments. A symbiotic relationship is established between bacterial flora and their host environment. The bacteria stimulate vascularization and development of intestinal villi and stimulate epithelial cells to secrete bacterial nutrients. The bacteria generate simplified carbohydrates, amino acids, and vitamins.
Not all bacteria found in the GI tract are harmless. One of the occupants is the Gram-positive bacterium Enterococcus faecalis. This is an opportunistic pathogen that occupies a variety of ecological niches including soil, sewage, and water. Inside the GI tract, this bacterium causes uninary tract infections and is highly resistant to vancomycin, the preferred antibiotic for treating gram-positive bacterial infections. The propensity for this bacterium to harm its host is tied to genetic material acquired through horizontal gene transfer (HGT) from other bacteria. More than 25% of its genome consists of DNA acquired from other species.
411

412 17. Cell Regulation in Bacteria
17.1Cell Regulation in Bacteria Occurs Primarily at Transcription Level
Messenger RNAs are rapidly turned over in bacteria. Their half-lives are far shorter than eukaryotic mRNAs. Whereas eukaryotic mRNAs have half-lives on the order of an hour, bacterial mRNA are degraded within a few minutes. Since the steady state concentration of any molecule is directly proportional to its half-life, the rapid degradation of selected molecules ensures that the bacterium can respond rapidly to changing environmental conditions. The rapid degradation of proteins is coupled to an equally rapid capability for synthesizing new proteins.
The machinery for regulating gene expression in bacteria is highly streamlined compared to that for eukaryotes. Bacterial DNA is not wrapped in histones, and chromatin-modifying enzymes are not required. Their genes do not possess introns, and a splicing stage is not needed. There are few noncoding regions, and the fraction of DNA that codes for proteins ranges from 87 to 95%. Transcription and translation are contiguous, and the two processes take place at the same time in the same place. In many species extra-chromosomal DNA is present. It is organized into one or more small circular plasmids containing collections of genes that perform specialized functions.
Bacteria such as Streptomyces coelicolor, Escherichia coli, Bacillus subtilis and Caulobacter crescentus possess genomes comparable in size to the unicellular yeast genome, and some, for example, Calothrix, have genomes approaching Drosophila in size. These bacterial species possess a sophisticated control layer that enables them to alter their metabolism, morphology, and lifestyle. Some can execute developmental programs that produce differentiated progeny and make possible colonial behavior. The first part of this chapter is devoted to an examination of gene regulation in bacteria, the counterpart to the discussions in the last chapter dealing with gene regulation in eukaryotes. The second part of this chapter deals with the more sophisticated forms of cell regulation in bacteria. Special attention is given to how some of these modes help cause disease in humans while other modes promote beneficial symbiotic relationships.
17.2 Transcription Is Initiated by RNAP Holoenzymes
Transcription in bacteria is carried out by RNA polymerase (RNAP) holoenzymes. Whereas eukaryotes possess three kinds of RNA polymerases, bacteria utilize a single kind. The bacterial RNAP molecule is composed of a large and rather nonspecific core RNAP unit and a small regulatory and targeting subunit, the sigma factor (Table 17.1).
The core unit is composed of four subunits—a pair of alpha subunits, and two larger subunits, b and b¢, responsible for the enzymatic activities of the
17.2 Transcription Is Initiated by RNAP Holoenzymes |
413 |
|||
TABLE 17.1. Components of the E. coli RNA poly- |
|
|||
merase holoenzyme. |
|
|
|
|
Subunit |
Molecular weight (kDa) |
Function |
|
|
a |
36.5 |
Targeting and assembly |
|
|
b, b¢ |
150.6, 155.2 |
Catalytic activities |
|
|
s |
20 to 70 |
Promoter recognition |
|
|
|
|
|
|
|
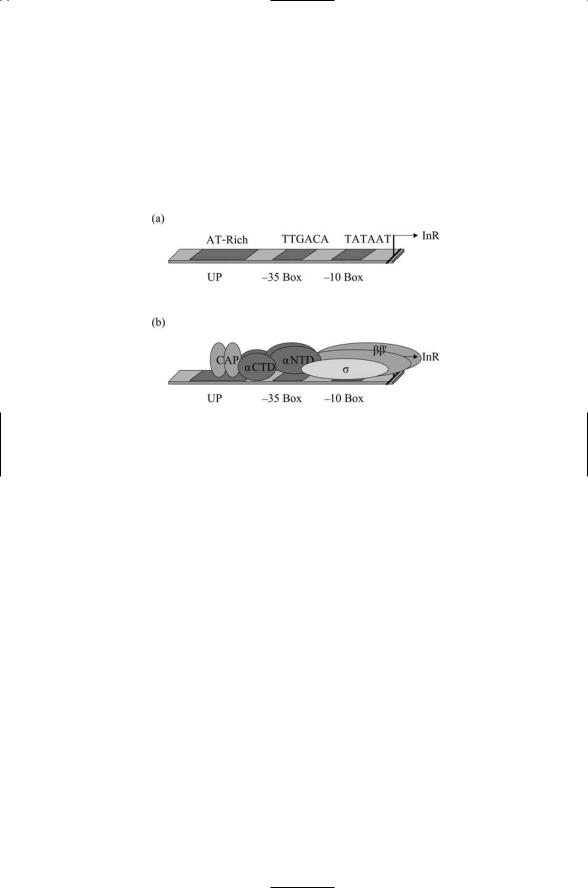
FIGURE 17.1. Organization of the bacterial promoter: (a) The primary DNA binding regions are shown together with the transcription start site, InR. (b) Arrangement of the subunits of RNA polymerase together with a catabolite activator protein (CAP) transcriptional activator dimer.
polymerase. The alpha subunit contains two domains connected by a flexible hinge. As shown in Figure 17.1b, each alpha subunit has an N-terminal domain (aNTD) that interfaces the subunit with the beta subunits, and a
C-terminal domain (aCTD) that mediates interactions with the DNA promoter and with regulatory proteins. The core, written symbolically as a2bb¢, has a total mass of about 380 kDa. There are several different kinds of sigma factors, and their masses vary from 20 to 70 kDa. The sigma factors are DNA sequence-specific binding proteins that recognize and bind the two specific promoter sites listed in Table 17.2. The bacterial transcription-competent RNAP holoenzyme is thus of the form a2bb¢s.
The association of a sigma factor and the core RNAP unit is transient. Shortly after initiation of transcription, the sigma factor dissociates from the core unit, which proceeds with transcript elongation. This dissociation makes possible the attachment of a different sigma factor subsequent to termination of the transcription event. If alternative sigma factors are present in sufficient numbers, they will out-compete the initial sigma factor for binding to the core unit. The core is thus reprogrammed to bind to an alternative promoter site and transcribe a different set of genes.

414 17. Cell Regulation in Bacteria
TABLE 17.2. Bacterial s70 promoter architecture.
Regulatory region |
Consensus sequence |
Binding |
-10 box |
5¢-TATAAT-3¢ |
s |
-35 box |
5¢-TTGACA-3¢ |
s, TFs |
UP element |
AT-rich tracts |
aCTD, TFs |
|
|
|
17.3Sigma Factors Bind to Regulatory Sequences in Promoters
DNA promoters that bind bacterial RNAP subunits and transcription factors contain a pair of core regulatory sequences separated by a spacer region. The regulatory sequence nearest the transcription (+1) start site is the -10 box, and it is centered 10 bp immediately upstream of that site. This region is separated by a spacer from the -35 box, the second core regulatory region centered 35 bp upstream. The amount of agreement between -10 and -35 box sequences and the consensus sequences has a regulatory role. Genes are strongly expressed at sites where the agreement is good and are weakly transcribed at sites where the agreement is not optimal. In many promoters, there is a third regulatory site located further upstream. Referred to as the upstream (UP) sequence its precise location varies from promoter to promoter (Figure 17.1a).
Different sigma factors recognize different promoter sequences. The consensus sequences listed in the second column of Table 17.2 are recognized only by primary sigma factor (i.e., by s70). Each of the groups of alternative sigma factors listed in Table 17.3 has its own characteristic -10 and -35 consensus sequences.There are two families of sigma factors, s70 and s54.
Members of the s54 differ from those in the s70 in several ways. One of the ways they differ is that instead of recognizing sequences -10 and -35 bp upstream of the start site they recognize sequences centered -12 and -24 bp upstream.
17.4Bacteria Utilize Sigma Factors to Make Major Changes in Gene Expression
Bacteria such as E. coli and B. subtilis express several kinds of sigma factors. The primary sigma factors, s70 in E. coli and sA in B. subtilis, regulate genes active all the time that perform housekeeping functions and support normal vegetative growth. This latter term denotes situations where nutrients are plentiful. Rapid cell growth and cell division typically produces exponential increases in the bacterial population. A second sigma factor is utilized in situations where exponential growth is not supported. The

17.4 Bacteria Utilize Sigma Factors |
415 |
TABLE 17.3. Sigma factors of E. coli and B. subtilis.
Sigma factor |
Bacteria and designation |
Control function |
Primary |
E. coli s70, B. subtilis sA |
Housekeeping, vegetative |
|
E. coli sS (s38) |
growth |
|
Stationary phase |
|
Heat shock |
E. coli sH (s32) |
Stress response genes |
|
E. coli sE (s24) |
Stress response genes |
|
B. subtilis sB (s37) |
Stress response genes |
Flagellar and chemotactic |
E. coli sF (s28), |
Genes for motility and |
|
B. subtilis sD |
chemotaxis |
Sporulation |
B. subtilis sH |
Postexponential, |
|
|
competence and early |
|
B. subtilis sE |
sporulation genes |
|
Early mother cell genes |
|
|
B. subtilis sF |
Early forespore genes |
|
B. subtilis sG |
Late forespore genes |
|
B. subtilis sK |
Late mother cell genes |
Extracytoplasmic function (ECF) |
E. coli sFecI (s19), |
Genes for iron uptake, |
|
B. subtilis sV, sZ |
antibiotic production, |
|
|
virulence factors, outer |
s54 family |
E. coli sN (s54), |
membrane proteins |
Nitrogen fixation, genes for |
||
|
B. subtilis sL |
degradative enzymes |
|
|
|
bacteria modify many of their internal activities in these cases using the stationary phase sigma factor sS. This sigma factor stimulates physiological changes needed in the presence of nutrient deprivation and toxic chemicals.
Many stressful conditions have to be dealt with by bacteria. Some conditions, like temperature extremes, osmotic stresses, and pH effects, are related to the chemical and physical environment. These stresses are dealt with by stress response sigma factors. High temperature is a representative example of a stressful condition. When high temperatures are encountered bacteria such as E. coli and B. subtilis use sH and sB to trigger the synthesis of heat shock proteins. These stress response proteins refold proteins that have become partially unfolded due to thermal effects, and if the proteins cannot be refolded tag them for destruction. Sporulation and extracytoplasmic function (ECF) sigma factors are two more families of sigma factors involved in controlling specific stress responses. The sporulation sigma factors control the development of spores in B. subtilis in response to starvation conditions. The ECF sigma factors control a diverse set of responses (for example, iron uptake, heavy metal resistance, antibiotic production, and induction of virulence factors) mostly involving the remodeling of the transport machinery located in bacterial membranes. Finally, some members of the s54 family are involved in control of nitrogen metabolism while others are involved in regulating stress responses.

416 17. Cell Regulation in Bacteria
17.5 Mechanism of Bacterial Transcription Factors
Most bacterial transcription factors contain a helix-turn-helix (HTH) motif consisting of about 20 amino acid residues that binds DNA and is part of a larger DNA binding domain. The DNA binding domain often has additional sites that contact DNA and aid in recognition. The transcription factors bind to regulatory sequences in the promoters as dimers or as higher order oligomers. Besides contacting the DNA, transcription factors also contact one or more subunits of the RNAP holozenzme. Most often they contact s and/or aCTD. However, some contact b, b¢, or aNTD. Promoters often contain sites for attachment of multiple transcription factors, and by this means integrate a variety of environmental signals into the transcription decision.
Bacterial TFs can be grouped into families according to characteristic DNA sequences present in family members and related physiological functions. Members of more than a dozen families of transcription factors are present in environmental bacteria such as Escherichia coli, Bacillus subtilis,
Vibrio cholerae, and Pseudomonas aeruginosa. A short list of the most prevalent families present in these organisms is presented in Table 17.4. As discussed in Chapter 1, the size of the control layer contracts rapidly when the genome sizes are reduced. Minimalist bacteria such as Mycoplasma genitalium have only a few transcription factors, or none at all. Social, differentiating bacteria, which will be discussed later in this chapter, possess genomes that are even larger than those of the environmental bacteria. These organisms may well possess families of transcription factors needed for differentiation that are absent in Table 17.4.
Bacterial transcription factors are HTH DNA binding proteins that integrate signals, and fine tune and regulate transcription in response to changes
TABLE 17.4. Transcription factors, their actions, and typical cellular roles.
Family |
Actions |
Cellular role |
AraC/XylS |
Activator |
Virulence, sugar metabolism |
ArsR |
Repressor |
Heavy metal resistance |
AsnC |
Dual |
Amino acid biosynthesis |
Crp |
Dual |
Global responses |
DeoR |
Repressor |
Sugar metabolism |
GntR |
Repressor |
Carbon metabolism |
LacI/GalR |
Repressor |
Carbon uptake |
LuxR |
Activator |
Biosynthesis, glycerol metabolism |
LysR |
Dual |
Amino acid biosynthesis |
MarR |
Repressor |
Antibiotic resistance |
MerR/SoxR |
Dual |
Heavy metal resistance |
TetR/AcrR |
Repressor |
Antibiotic resistance |
|
|
|

17.6 Many TFs Function as Response Regulators |
417 |
in their environment. First and foremost, the transcription factors regulate metabolic activities. Sugar metabolism, carbon uptake and metabolism, and amino acid biosynthesis are heavily represented in the list of cellular roles in Table 17.4. The second main function of the transcription factors is to regulate physiological responses to potentially harmful substances in the environment. Noteworthy in the table are transcription factors for arsenic resistance (ArsR), for mercury, copper, zinc, and cobalt resistance (MerR), and for tetracycline and other antibiotics resistance (TetR).
17.6 Many TFs Function as Response Regulators
Many transcription factors function as response regulators in two-component signal transduction systems. Two-component signal transduction systems are the main devices used by bacteria to sense their environments. The bacterial chemotactic system discussed in Chapter 6 is the prototype example of a two-component system. It is used to sense nutrients and noxious compounds in the local environment and relay these signals to a flagellar motor so that a bacterium can swim towards nutrients and away from harmful substances. More commonly, two-component systems are used to relay environmental information directly to sites on chromosomes and plasmids that regulate transcription.
Recall that two-component systems consist of a sensor module and a response regulator module. In two-component systems that regulate transcription the sensor module is a single transmembrane protein and the response regulator is a single cytoplasmic protein. The sensor, a histidine protein kinase, contains an N-terminal input unit and a C-terminal transmitter unit. The input unit responds to environmental stimuli by stimulating the autophosphorylation of the transmitter on a specific histidine residue. The high energy phosphoryl group is then transferred to the second component of the system, the response regulator. The response regulator contains an N-terminal regulatory domain and a C-terminal DNA binding domain. The N-terminal domain catalyzes the transfer of the phosphoryl group to a conserved aspartate residue in its own domain. The C-terminal output unit possesses an HTH DNA-binding domain that functions as a transcription factor. The output unit is activated in response to phosphorylation state-dependent conformational changes in the regulatory domain.
The second column of the Table 17.4 describes the regulatory function (action) of the transcription factors. Some TFs are activators: They are positive regulators that stimulate transcription at higher frequencies and/ or for greater durations. Other transcription factors are repressors: They are negative regulators that block or mechanically inhibit transcription. Still other transcription factors can act either as activators or as repressors: They are dual regulators and their specific actions will vary with cellular context.

418 17. Cell Regulation in Bacteria
Transcription activators bind to sites upstream of the -35 box. Some bind in the vicinity of the UP element while others bind to sites further upstream. Many of these transcription activators exert their influences by binding to aCTD. Because of its flexibility, aCTD can bind to activators over distances from -35 to -100 bp. Transcription factors that bind to distant upstream sites are often referred to as transcription enhancers. The DNA molecule itself has a certain intrinsic flexibility. Sequences such as A-T tracts are especially bendable. This flexibility plus bending brought on by protein binding enables activators to come into physical contact with subunits of the holoenzyme. Transcription repressors bind to sites close to the transcription start site. These proteins prevent the holoenzyme from initiating transcription either by sterically inhibiting access or by binding the holoenzyme so tightly that it cannot release and start transcription.
17.7Organization of Protein-Encoding Regions and Their Regulatory Sequences
Bacterial DNA is organized into operons. Each operon contains DNA sequences that encode proteins and the upstream sequences for attachment of the RNA holoenzyme and regulatory proteins (Figure 17.2). Regulatory information is conveyed in several ways: It is imparted through the promoter recognition sequences that provide attachment sites for specific sigma factors: it is conveyed by degree of agreement between the promoter sequences and the consensus sequence, and by the length of the intervening spacer sequences, both influencing the strength of the transcription; and it is supplied by the presence of binding sites for transcription activators and repressors.
In contrast to transcription in eukaryotes, several genes can be transcribed from each promoter into an mRNA molecule. The protein-coding sequences follow one another and the protein-coding region terminates with a characteristic stop sequence that mediates the release of the polymerase from the DNA. Multiple gene transcription makes it possible for a promoter to regulate its own activity by implementing a simple negative feedback loop. Two kinds of proteins—structural and regulatory—can be
FIGURE 17.2. Bacterial operon: Shown are five genes, labeled A through E, controlled by a single gene regulatory region partitioned into repressor (O), core promoter (P), and enhancer (E) sections.

17.8 The Lac Operon Helps Control Metabolism in E. coli |
419 |
expressed at the same time. In negative feedback, regulatory proteins shut down transcription from their own promoters, thus providing a mechanism for automatically terminating transcription an appropriate time after the onset of transcription as determined by the threshold concentration. In many instances the regulatory proteins control the assembly of the cotranscribed structural proteins into membrane-bound structures.
Cassettes have the following properties. They are sets of operons that encode proteins performing a set of related functions. They are usually found on plasmids and enable the bacteria to respond to hostile conditions. The operons are positioned in the vicinity of one another, and because of their location on plasmids can be transferred from one bacterium to another. Some cassettes encode proteins involved in either antibiotic production or antibiotic resistance. Others, called virulence cassettes or pathogenicity islands, encode virulence factors such as toxins and proteins required for erection of morphological structures involved in the toxins’ secretion and delivery to targets.
Regulons are sets of operons located at well-separated loci along the chromosome; they encode proteins involved in a common physiological response. Many regulons are organized by sigma factors. By binding to multiple sites along the chromosome they can either upregulate or downregulate 100 or more proteins. Examples of sigma factor associated regulons include the E. coli sS stationary phase regulon, the E. coli s32 heat shock regulon, and the B. subtilis sB stress response regulon.
17.8The Lac Operon Helps Control Metabolism in
E. coli
Bacteria optimize their metabolism to take maximal advantage of their environments. Bacteria such as E. coli possess a broad range of metabolic strategies. They are able to metabolize a variety of sugars including glucose, lactose, and maltose, and make best use of the nutrients available in their environment. E. coli is the most studied bacteria and serves as a model system for how these organisms rapidly adjust their metabolism to environmental conditions. The genes involved in regulating whether glucose or lactose is metabolized form the lac operon. They are listed in Table 17.5, and their arrangement in the lac operon is illustrated in Figure 17.3.
TABLE 17.5. Genes belonging to the E. coli lac operon.
Gene |
Function |
Description |
LacI |
Repressor |
Binds to transcription start site and part of the |
|
|
promoter, inhibiting RNAP |
LacZ |
b-galactosidase |
Hydrolyzes lactose to glucose plus galactose |
LacY |
b-galactoside permease |
Transports lactose into the cell |
LacA |
b-galactoside transacetylase |
Nonessential enzyme |
|
|
|
420 17. Cell Regulation in Bacteria
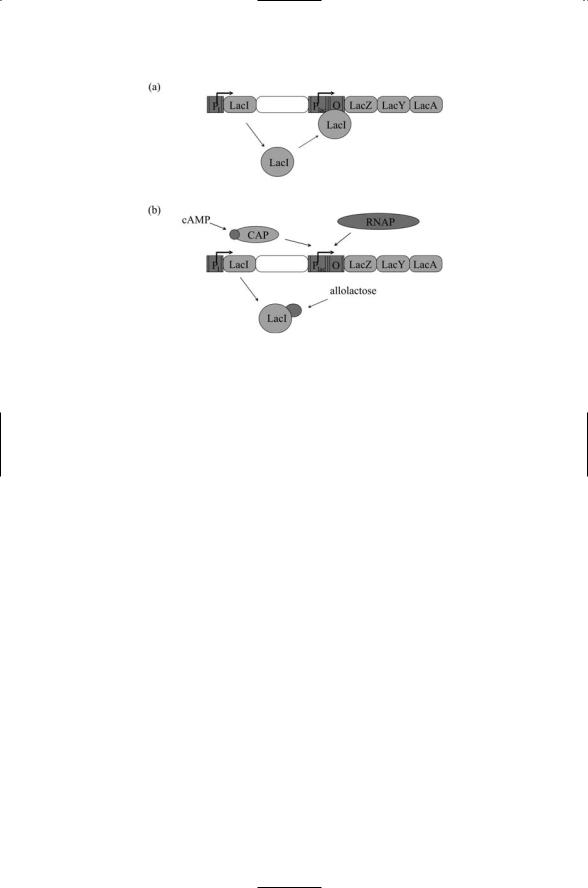
FIGURE 17.3. Organization of the lac operon: (a) Constitutive expression of the lac repressor protein (LacI) blocks transcription from the lac promoter when it binds to the operator. (b) Binding of LacI is blocked by allolactose allowing cAMP-bound CAP proteins to bind and stimulate transcription by the NRA polymerase (RNAP).
Glucose is the favored sugar and if it is plentiful the expression of proteins needed to metabolize alternative nutrients is inhibited. The LacI repressor is constitutively expressed and under plentiful glucose conditions binds downstream of the -10 and -35 sites and blocks RNAP from initiating transcription. When lactose is present in the environment, some of it will enter the cell and, in the form allolactose, will bind to the LacI repressor. The lactose binding induces structural changes in LacI and the repressor is no longer able to block RNAP.
The lac promoter is a weak one. The sequences at the -10 and -35 sites are not optimal for RNAP binding and the polymerase needs the binding activities of an activator, the catabolite activator protein (CAP), to efficiently transcribe the structural genes in the operon. CAP forms a complex with cAMP. When glucose levels are high the cAMP levels are low and the complex does not form. When glucose is absent, the cAMP levels are high enough for the CAP/cAMP complexes to form. The complexes bind at a site upstream of the RNAP binding site (CAP by itself does not), where they help RNAP to bind and start transcribing the genes. In sum, under the combined actions of the LacI (negative regulation) and CAP (positive regulation), the lac genes are transcribed when lactose is present and glucose is absent (Figure 17.3).
The structure of LacI is well suited for controlled interactions with DNA. As shown in Figure 17.4, two of its domains, the so-called Headpiece and
