
Molecular and Cellular Signaling - Martin Beckerman
.pdf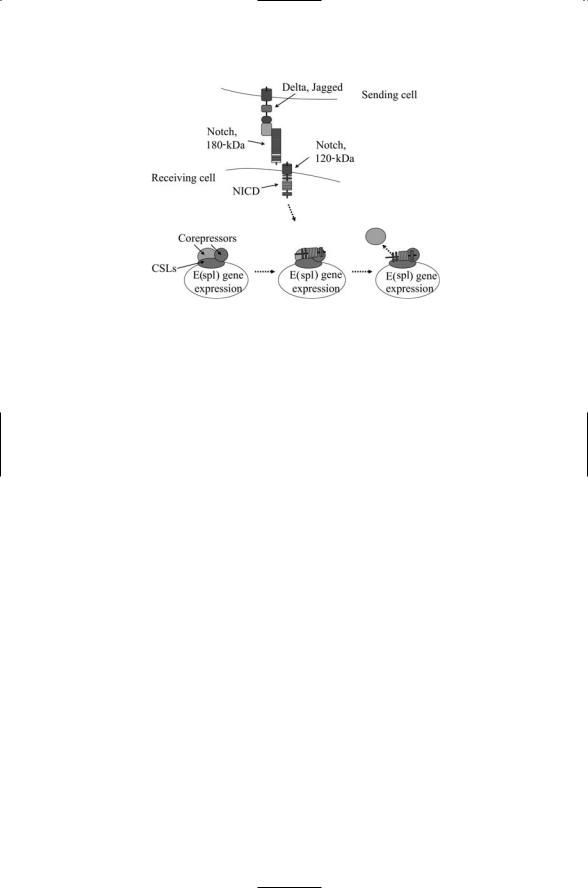
308 13. Cell Fate and Polarity
FIGURE 13.2. Signaling through the Notch pathway: The sending cell expresses the Notch ligand Delta or Jagged, while the receiving cell expresses the Notch receptor. The extracellular chain of Notch binds the ligand and in response the transmembrane segment is cleaved at several places to form the NICD. The NICD, together with Su(H), translocates to the nucleus where they interact with several other proteins to activate gene transcription. As depicted in the figure, they may promote gene transcription by displacing corepressors.
ing along this fate pathway by promoting either a different pathway or by maintaining the cells in an uncommitted state. The general mechanism for restricting cell fate decisions is called lateral inhibition because a cell adopting a particular cell fate inhibits its neighbors from adopting the same fate.
A key factor that promotes cell fate decisions is the use of a positive feedback loop where small stochastic differences in expression levels are amplified and drive the decision. Lateral inhibition works in the following manner (Figure 13.3). Cells express both Notch and Delta on their surface. Notch receptors on one cell bind to Delta ligands on the other. When a cell’s Notch receptor binds a Delta ligand on an opposing cell it develops a diminished capacity for expressing Delta ligands on its own surface. Cells expressing Notch more strongly will receive a stronger inhibitory signal and inhibit its neighbor less strongly. This generates a feedback loop that amplifies initially small stochastic differences in receptor expression.
13.3Proteolytic Processing of Key Signaling Elements
Proteolytic processing plays a key role in many signaling pathways. Notch is cleaved after ligand binding, and the cytosolic fragment translocates to the nucleus where it functions as a transcription factor. Thus, proteolytic

13.3 Proteolytic Processing of Key Signaling Elements |
309 |
FIGURE 13.3. Lateral inhibition through cell-to-cell signaling and positive feedback:
(a) Because of stochastic fluctuations there are more Notch receptors on the B cell than on the A cell and, other things being equal (i.e., the density of Delta ligands on the two cells being the same), signaling into the B cell is stronger than in the A cell. (b) In the B cell, Notch signal transduction upregulates Notch receptor expression and reduces Delta ligand expression. On the A cell the strength of Notch signaling fails to compensate for the decay of Notch receptors over time. Thus on the B cell Delta ligand expression goes down while on the A cell Notch receptor expression declines. (c) This tendency is amplified over time leading to an A cell expressing Delta ligands and a B cell expressing the Notch receptors.
processing enables a single gene product to function first as a receptor and then as a transcription factor. As noted earlier proteolytic processing of the 300-kDa precursor takes place in a trans-Golgi network resulting in formation of the two-chain receptor.
Proteolytic processing plays key role in signaling through the Hedgehog and Wnt pathways, too. As will be seen shortly in Sections 13.6 and 13.7 these processes are, in turn, regulated by protein phosphorylation. In these pathways, these two forms of posttranslational modification work together to activate elements of the signaling pathways located within the cell, in the cytosol, in response to ligand binding at the plasma membrane.
Several families of enzymes participate in cleaving the transmembrane form of the Notch receptor leading to release of the NICD. One of the families of proteolytic enzymes is the Presenilins. These enzymes have been the subjects of intensive study in the past few years due to their similar involve-

310 13. Cell Fate and Polarity
FIGURE 13.4. Components of the g-secretase: (a) Presenilin—The small squares denote the location of a pair of aspartate residues crucial for the catalytic activities of presenilin; (b) Nicastrin—Ovals located on extracellular part of the chain denote essential glycosylation sites. (c) Aph-1. (d) Pen-2.
ment in cleaving the amyloid precursor protein (APP) leading to the release of the Ab amyloid protein, an important step in the onset of Alzheimer’s disease. Like the Notch process depicted in Figure 13.1 the APP undergoes several cleavages. These are mediated by enzymatic complexes referred to as a-secretases, b-secretases, and g-secretases. The Presenilins are part of the g-secretase complex, which has three other members—Nicastrin, Aph- 1 and Pen-2. As illustrated in Figure 13.4 the Presenilins are 8-pass transmembrane (TM) proteins proteolytically cleaved into two chains. A pair of aspartate residues positioned in opposition to one another is crucial for Presenilin’s g-secretase actions. Notch and APP pass through the gap formed by the TM segments containing the aspartates, and are cleaved. The Presenilins carry out their catalytic activities together with the single-pass Nicrastrin protein, the 7-pass Aph-1 protein, and the 2-pass Pen-2 protein, which help to assemble and stabilize the complex.
The APP is cleaved twice, one in its extracellular domain just outside the membrane and the other near the middle of the intermembrane region. In the first step, the APP proteins are cleaved at one of two alternative extracellular locations termed the a- and b-sites. The g-secretases are responsible for subsequent cleavages within the transmembrane segment. In the case of cleavage at b-sites, but not a-sites, the products are 40and 42-amino acid residue forms of the Ab amyloid protein. The 42-residue form, favored by the mutated Presenilins, is especially prone to form clumps due to exposure of hydrophobic patches, as was discussed in Chapter 5. The a- secretases and b-secretases responsible for cleaving the APPs at the a- and b-sites are members of two other families of proteases. One or more members of the “a disintegrin and metalloprotease” (ADAM) family are the a-secretases, while aspartyl proteases belonging to the membraneassociated aspartyl protease (memapsin) family serve as the b-secretases.
The ADAM enzymes are members of a large group of multidomain enzymes that possess not only metalloproteinase domains, but also integrinbinding regions and a cytoplasmic domain that bind a variety of signaling

13.4 Three Components of TGF-b Signaling |
311 |
proteins. Members of the ADAM family of proteases degrade collagens and other extracellular matrix proteins. This activity is particularly important during embryonic development. During that time the ECM has to be continuously remodeled to accommodate new growth and emerging organs and appendages. The ADAM proteins also promote the creation of soluble signaling proteins from membrane-bound forms in another developmentally important process called ectodomain shedding. In this process, membrane-bound proteins are cleaved at sites just outside the plasma membrane, thereby converting ligands and receptors that signal in a short range, juxtacrine manner into soluble signaling proteins that can signal in a longer-range, paracrine manner. One of the ADAM family members, a protein called tumor necrosis factor-a converting enzyme (TACE) can cleave the TNF-a ligand, as its name suggests, and functions as the a- secretase for Notch and the APP perhaps along with another protein called ADAM10 in mammals and Kuzbanian in Drosophila. Lastly, degradation of ECM molecules is necessary for cell migration. The upregulation of metalloproteases (metalloproteinases) accompanies the downregulation of cell adhesion molecules, increased angiogenesis, and increased motility in the progression of cancer to metastasis.
13.4 Three Components of TGF-b Signaling
The TGF-b signaling pathway has three main components: ligands, receptors and Smad signal transducers. The transforming growth factor-b pathway, like the Notch pathway, has many different effects on cell fate depending on the stage of development. Among the processes triggered through this pathway are patterning, wound healing, bone formation, ECM production, and homeostasis. There are three core components in a TGF-b pathway—a ligand, a cell surface receptor complex, and a set of cytoplasmic signal transducers called Smads. The TGF-b pathway is named for the TGF-b subfamily of diffusible polypeptide ligands. Other prominent ligands involved in signaling though the TGF-b pathway are the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), members of the Activin subfamily, and Nodal.
TGF-bs, BMPs, GDFs, and related ligands signal through receptors belonging to the TGF-b receptor family. These receptors are transmembrane glycoproteins possessing cytoplasmic domains that phosphorylate serine and threonine residues of target proteins. Thus they belong to the family of receptor serine/threonine kinases. A sequence of events involving one or more members of two distinct TGF-b receptor groups is required in order to signal across the plasma membrane. The two receptors groups are designated Type I and Type II receptors, abbreviated as TbRI and TbRII. Members of each group possess an extracellular ligand-binding domain and a cytoplasmic kinase domain. Type I receptors have an additional cytoplas-
312 13. Cell Fate and Polarity
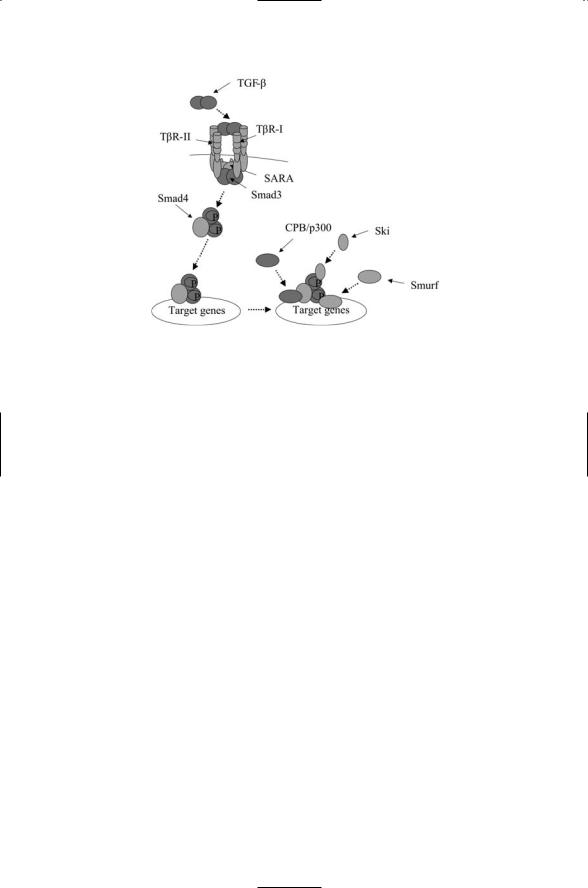
FIGURE 13.5. Signaling through the TGF-b pathway: Binding of a ligand (TGF-b) dimer to a TbRII-TbRI receptor complex leads to phosphorylation of serine/threonine residues in the GS region of the TbRIs resulting in the activation of the kinase domain. The activated receptors phosphorylate the Smad3 proteins leading to the latter’s dissocation from the SARA anchors. They form a heterotrimeric complex with Smad4, and in that form translocate to the nucleus where they form a scaffold for assembly of transcriptional activators and cofactors such as CPB/p300 and Ski to induce the transcription of target genes.
mic domain referred to as a GS domain that functions as a key regulatory region.
Two different mechanisms of ligand-receptor binding are utilized by
TGF-b receptors. BMP ligands have a high affinity for Type I receptors and a low one for Type IIs. Once BMP ligands bind the Type Is, these receptors have a high affinity for Type II receptors. The assembly setups are, as follows. First, a complex is formed containing a BMP ligand dimer bound to a Type I receptor dimer. This assembly then binds to two Type II receptors to make a receptor foursome, as shown in Figure 13.5. TGF-b and Activin utilize a different assembly method. These ligands prefer to bind to Type II receptors. Their first step is the binding of ligands to Type II receptor molecules. These binding events serve to recruit Type I receptor molecules to the assemblage, resulting again in the formation of a complex consisting of a ligand dimer bound to a receptor foursome. As a consequence of either series of binding events the cytoplasmic domains of the Type II and Type I proteins are brought into close proximity and stabilized.
The next step in transducing the signal into the cell is transphosphorylation of serine and threonine residues in the GS domains of the Type I receptors, assisted by the Type II receptors. This step creates docking sites for a

13.5 Smad Proteins Convey TGF-b Signals into the Nucleus |
313 |
number of different adapter proteins. One of these, the Smad anchor for receptor activation (SARA), couples Smad2/3 to the activated receptor complex. The SARA protein possesses a lipid-binding FYVE domain that enables it to tether to the plasma membrane. Two other domains permit it to bind simultaneously to the receptor and to the Smad protein. Once the Smad proteins have been recruited to the receptor complex they are phosphorylated, triggering their release into the cytosol (Figure 13.5).
13.5Smad Proteins Convey TGF-b Signals into the Nucleus
The Smad proteins convey messages from the cell surface to the nucleus. The name “Smad” is a contraction of the names of the first two Smad-type proteins to be discovered—the C. elegans Sma protein and the Drosophila mothers against dpp, or Mad, protein. There are at least eight known Smads, designated Smad1 through Smad8, and they fall into three categories (Table 13.2). Five of the Smads are receptor-activated and they are called R-Smads. Smad4 is required for signaling through all pathways and is called a common Smad, or co-Smad. The other two Smads, Smad6 and Smad7 are inhibitory. They turn off signaling and form an anti-Smad grouping. Smad 6 inhibits signaling through the BMP branch while Smad7 inhibits signaling through all of the branches of the TGF-b signaling pathway.
Smad proteins contain a pair of globular signaling domains connected by a linker region. The N-terminal Mad Homology-1 (MH1) and C-terminal
TABLE 13.2. The TGF-b signaling pathway: As indicated in the table, the TGF-b pathway branches at the Type I receptors (Type I Rs) level into an Activin-like branch and a BMP-like branch. Abbreviations—Activin receptor-like kinase (ALK); bone morphogenetic protein (BMP); transforming growth factor (TGF); receptor (R); inhibitory (I).
|
Activin |
TGF-b |
TGF-b |
BMP |
Ligands |
Activins |
TGF-bs |
TGF-bs |
BMPs |
Type II Rs |
ActII/IIB |
TbRII |
TbRII |
BMPRII, |
|
|
|
|
ActRII/IIB, |
|
|
|
|
MISRII |
Type I Rs |
ALK4 |
ALK5 |
ALK1 |
ALK3, ALK6, |
|
|
|
|
ALK2 |
R-Smad |
Smad2, Smad3 |
Smad2, Smad3 |
Smad1, Smad5, |
Smad1, Smad5, |
|
|
|
Smad8 |
Smad8 |
Co-Smad |
Smad4 |
Smad4 |
Smad4 |
Smad4 |
I-Smad |
Smad7 |
Smad7 |
Smad6, Smad7 |
Smad6, Smad7 |
|
|
|
|
|

314 13. Cell Fate and Polarity
Mad Homology-2 (MH2) domains mediate protein-DNA (MH1) and protein-protein (MH2) interactions. In addition, the R-Smad proteins contain a phosphorylation site near their C-terminal. Once activated by TbRI, the R-Smads translocate to the nucleus. On the way to the nucleus the R-Smads associate with the co-Smad (Smad4) as illustrated in Figure 13.5 for the case of Smad3.
The specific cell fate decision arrived at through the TGF-b pathway depends on context. The steps outlined above are general and are neither cell-specific or developmental stage-dependent. Partner and accessory signaling molecules that combine with the Smads to form the active transcriptional complexes supply the contextural information. Coactivators, corepressors and other partners provide cell type and developmental stage inputs, and they fix parameters such as the duration of transcription. A few of these cofactors are presented in Figure 13.5. The p300 and CBP proteins are coactivators. A number of corepressors associate with the Smad proteins. Examples of these additional factors are the Sloan-Kettering Institute proto-oncogene (Ski), included in the figure, and the Ski-related novel gene N (SnoN) and the TG3-interacting factor (TGIF), not included.
Smads are regulated by ubiquitination. Recall that ubiquitination prepares substrate proteins for proteolytic destruction by the proteosome. It involves sequential operations by E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligase enzyme. As is the case for all four of the pathways discussed in this chapter, selective, regulated proteolysis is an important mechanism for controlling what signals are sent on to the nucleus. In the TGF-b signaling pathway, proteolytic regulation of the Smads takes place both in the cytosol and in the nucleus. Members of the Smurf family of E3 ligases are recruited to the Smad scaffolds. The WW domains of the Smurfs bind PPXY motifs in the linker region connecting the MH1 and MH2 domains of the Smads. Members of the UbcH5 family of E2s are present, as well. These operations are represented in
Figure 13.5 by the presence of a generic Smurf protein.
13.6Multiple Wnt Signaling Pathways Guide Embryonic Development
The Wnt pathway is named for the Wnt family of secreted glycoproteins, which function as morphogens during development in vertebrates and invertebrates. Wnt ligands are typically 350 to 400 amino acid residues in size and are characterized by a highly conserved cysteine-rich domain
(CRD), a pattern of 23 cysteine residues. A large number of Wnt proteins have been identified. There are at least 25 vertebrate Wnt genes, 7 Drosophila genes, and 5 C. elegans genes. The most prominent of the Drosophila Wnt family members is called Wingless (Wg), and is the homolog to the Wnt-1 protein found in humans. This ligand is responsible

13.6 Multiple Wnt Signaling Pathways Guide Embryonic Development |
315 |
FIGURE 13.6. Wnt signal receptors and transducers: (a) LRP coreceptor. (b) Frizzled receptor. (c) Strabismus co-receptor. The rectangle attached to the cytoplasmic C-terminal tail of the receptor denotes a PDZ binding domain. (d) Dishevelled adapter. Abbreviations—Dishevelled, Egl-10, and Pleckstrin (DEP); Dishevelled and axin (DIX). The DIX domain mediates attachment to the cytoskeleton, and the DEP domain promotes binding to the plasma membrane.
for a well-studied anterior/posterior decision that defines tissue polarity. (Note: Wnt is a contraction of wingless (Wg), and the murine, or mouse, homolog Int.)
Wnt ligands bind to Frizzled receptors. These receptors are named for the Drosophila frizzled gene involved in the tissue polarity decisions. Frizzled proteins have a large extracellular CRD responsible for ligand binding, a 7-pass (serpentine) transmembrane region and a short cytoplasmic C- terminal region. Several coreceptors operate in conjunction with the Frizzled receptors to transduce signals into the cell following ligand binding. The chain topology of Frizzled and two of its coreceptors, LRP and Strabismus, are shown in Figure 13.6. As shown in the figure, LRP is a single-pass receptor while Strabismus passes back and forth through the plasma membrane four times. The first cytoplasmic step following ligand binding in some Wnt pathways is the recruitment of an adapter protein, Dishevelled, to the plasma membrane. Coreceptors such as Strabismus exert their influences by interacting with this adapter protein.
The Wnt signaling pathway has several branches. One of these is referred to as the canonical or b-catenin pathway. Another is termed the planar cell polarity (PCP) pathway. As depicted in Figure 13.7, this pair of pathways splits when signals reach the cytoplasmic adapter protein Dishevelled. The decision of whether to activate the b-catenin or the planar polarity pathway is determined by interactions between the coreceptors and this adapter. The Strabismus coreceptor interacts with Dishevelled through their PDZ domains. It shifts the routing of the signals away from the b-catenin pathway and towards the PCP pathway by isolating the adapter from the Frizzed receptor, while the LRP coreceptor assists in signaling through the canonical pathway.
The canonical Wnt pathway leads through b-catenin. This protein is a transcription activator, but in the absence of Wnt signaling is prevented from performing this function. It forms a complex in the cytoplasm with
316 13. Cell Fate and Polarity
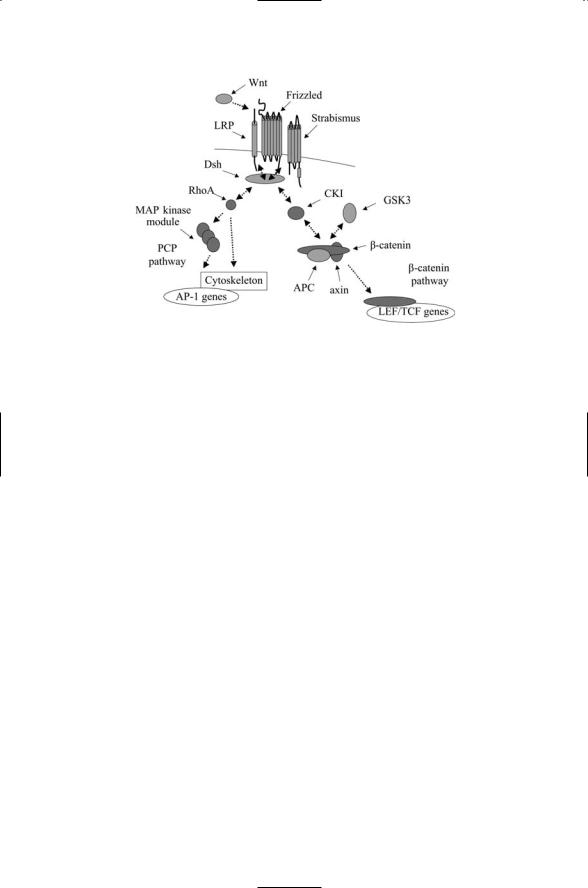
FIGURE 13.7. Signaling through the Wnt b-catenin and PCP pathways: Ligand binding activates either the canonical b-catenin pathway or the planar cell polarity pathway, depending on the actions at the Dishevelled switching point. In the canonical pathway, Dishevelled acts through the CKI kinase to influence the decision whether LEF/TCF gene transcription is repressed or activated. Alternatively, Dishevelled acts through the small GTPase protein RhoA and downstream MAP kinases such as JNK to promote cytoskeleton polarization and gene expression.
axin, the adenomatous polyposis coli (APC) protein, and two serine/threonine kinases—GSK3 and CKI. This module works in the following way. The kinases phosphorylate the other members of the complex. Phosphorylation of axin stabilizes it; phosphorylation of APC enhances its interactions with b-catenin. b-catenin cannot signal because phosphorylation tags it for proteolytic destruction. As a consequence of the phosphorylations, b-catenin does not translocate to the nucleus and stimulate transcription. Dishevelled acting through CKI destabilizes the complex, and leads to dephosphorylation of b-catenin. The number of mobile and untagged b-catenin molecules increases, and they are able to translocate to the nucleus and stimulate transcription of TCF/LEF genes (Figure 13.7).
The presence of two serine/threonine kinases in the complex leads to a two-step phosphorylation/degradation process. CKI acts first to phosphorylate substrate residues within the complex. This action “primes” the system for subsequent phosphorylation by GSK3, which is then followed by the recruitment of proteolytic elements to b-catenin. In more detail, CKI phosphorylates b-catenin at Ser45. This action enables GSK3 to phosphorylate b-catenin at three neighboring sites (Ser33/Ser37/Thr41). In the absence of the priming CKI phosphorylation, GSK3 cannot phosphorylate b-catenin at the aforementioned sites.

13.8 Hedgehog Signaling Role During Development |
317 |
13.7 Role of Noncanonical Wnt Pathway
The noncanonical Wnt pathway guides the development of planar cell polarity and convergent extension. Planar cell polarity (PCP) is the term used to describe coordinated patterns of polarization of cells lying within the planar epithelium or sheet. The two best-known examples of PCP are in the Drosophila wing and compound eye. In the wing, hairs produced by epithelial cells all point in the same direction, towards the distal tip. In the compound eye, oriented hexagonal arrays of photoreceptor cell clusters (ommatidia) are formed. In vertebrates such as frogs and fish, cells in a tissue shift about and change shape so that their distribution becomes narrower along one axis and longer about an axis perpendicular to the first one. These developmental rearrangements are referred to as convergent extension. They, too, are guided by signaling through the PCP pathway.
Signals sent through the planar cell polarity pathway influence the organization of the cytoskeleton and gene expression. Whereas signals were routed by Dishevelled to the CKI and GSK3 kinases and to the b-catenin module in the b-catenin pathway, they are routed by Dishevelled to the RhoA GTPase and then to JNK family of MAP kinases in the PCP pathway resulting in activation of c-Jun and AP-1 dependent transcription (Figure 13.7). A third route, which may actually be a branch of the PCP pathway since it appears to involve Dishevelled and Strabismus, utilizes G proteins, increased Ca2+ second messenger production, and activation of calciumdependent serine/threonine kinases and phosphatases such as CaMKII, PKC, and calcineurin.
One of the crucial steps in generating planar cell polarity is symmetry breaking—the creation of asymmetries in the cell population using components that are initially distributed uniformly about each cell. Several proteins working in conjunction with Strabismus and Dishevelled accomplish this task. One of these proteins is an adapter protein called Prickle. This protein binds to Strabismus and sequesters Dishevelled near the coreceptor and away from Frizzled. A feedback loop amplifies initial random and small differences in Prickle concentration to produce sharp asymmetries between cells that signal to the nucleus through the PCP pathway and those for which this pathway is shut down.
13.8 Hedgehog Signaling Role During Development
The fourth signaling pathway to be discussed in this chapter is the signaling pathway named for the Hedgehog (Hh) family of diffusible cell-to-cell signaling molecules. Members of the Hh family include Hedgehog in Drosophila and at least seven vertebrate Hedgehogs, the most prominent of which are Sonic hedgehog (Shh), Desert hedgehog (Dhh), and Indian hedgehog (Ihh). This family regulates a wide spectrum of developmental
