
Kluwer - Handbook of Biomedical Image Analysis Vol
.2.pdf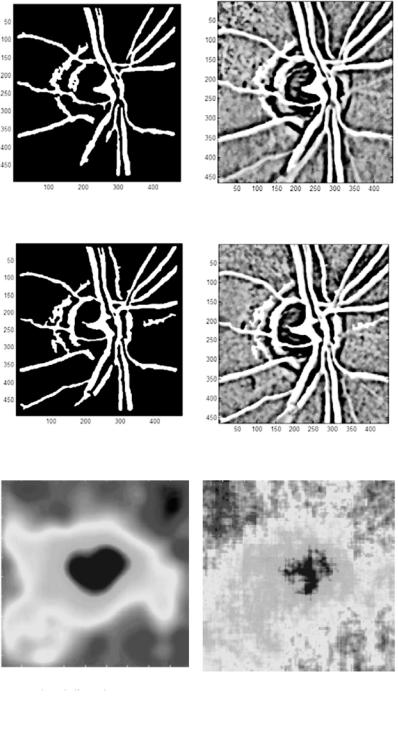
302 |
Yang and Mitra |
Segmented blood vessels (left) |
Feature map (left) |
Segmented blood vessels (right) |
Feature map (right) |
Interpolated disparity map Disparity map
(a) Disparity map with old segmentation technique
Figure 6.19: Disparity maps generated with and without DA feature extraction.
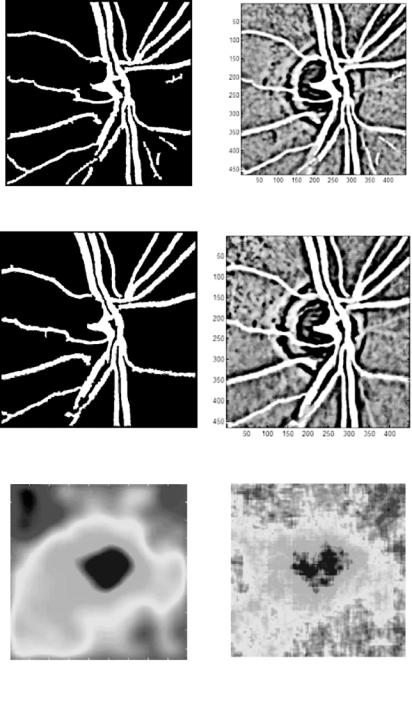
Statistical and Adaptive Approaches for Optimal Segmentation |
303 |
Segmented blood vessels (left) |
Feature map (left) |
Segmented blood vessels (right) |
Feature map (right) |
Interpolated disparity map |
Disparity map |
|
|
(b) New DA-based segmentation technique |
|
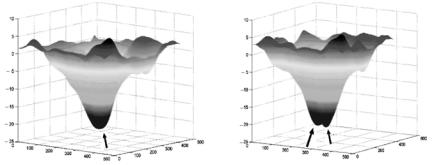
304 |
Yang and Mitra |
(c) Disparity map obtained from DA blood vessel segmentation
(d) Disparity map obtained from general edge detection for blood vessel segmentation
Figure 6.19: (cont.)
of cervical lesions. Automated image analysis is helpful in providing quantative lesion description thus monitoring of chronic lesions so that the onset of cervical cancer can be treated effectively.
A cervix image is a magnified color photograph of the cervix (illustrated in Fig. 6.20(a)). The acetowhite lesion area below the opening is marked by a trained physician, serving as a reference to other segmented images resulting from the algorithms. This image is taken with a regular high-resolution color camera, thus the most prominent problems preceding segmentation of the acetowhite lesion area from the rest are the reduction or removal of the glare and non-uniform illumination. Figure 6.20(b) shows the segmentation without illumination correction. All algorithms fail to recognize the lesion close to the cervix opening because the area is darker than other parts of the lesion, and vice versa for the section in the lower part of the cervigram, where the normal area is falsely classified as lesion. The glare on the top left of the lesion is also misclassified. Figures 6.20(c)–6.20(f) are the segmentation results from k-means, DA, and AFLC after illumination correction. The results are similar, with DA generating the closest partition to the manual segmentation.
6.5 Conclusions
From segmentation of the MR images, it can be observed that DA provides the best performance in terms of accuracy and stability among all discussed
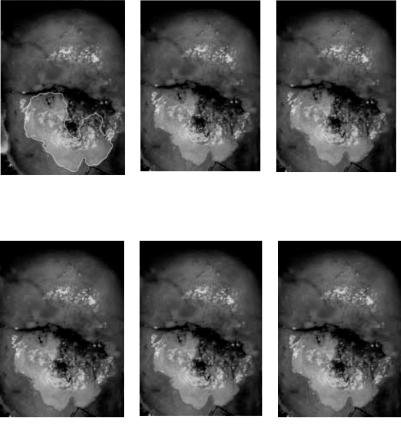
Statistical and Adaptive Approaches for Optimal Segmentation |
305 |
(a) Lesion |
marked by (b) Segmentation affected by |
(c) k-means-segmented |
physician |
nonuniform |
image (four clusters) |
|
illumination |
|
(d) DA-segmented lesion |
(e) AFLC-segmented lesion |
(f ) AFLC-segmented lesion |
(four clusters) |
(eleven clusters) |
(four clusters) |
Figure 6.20: Segmentation of cervical lesion.
clustering algorithms in several aspects. It is unsupervised; since theoretically it is designed to reach global minimum, the result is not biased by initialization. For the same type of image, the pseudotemperature reduction rate can be fixed, thus the segmentation process does not need parameter manipulation thereby yielding fully automated processing. Although the processing speed of DA is slower than other clustering algorithms, for small images such as the ones used above (217 × 181), the processing speed of DA is comparable to the other algorithms used. DA is also noise tolerant because of its statistical nature.
Both k-means and FCM, the well-known clustering algorithms, suffer from the initialization and local minimum problems. Cluster initialization is crucial in
306 |
Yang and Mitra |
yielding satisfactory results. When not initialized properly, a clustering algorithm might be trapped in a local minimum, failing to proceed to the correct cluster. Our experimentations show that with random initialization, both k-means and FCM fail to generate the lesion clusters in MRI MS segmentation. AFLC is an automated and adaptive improvement over k-means and FCM by incorporating neural leader clustering and FCM. The performance is improved; however, similar problems are still encountered. Initialization is eliminated by selecting the first incoming sample as initial centroid, therefore, the outcome is sampleorder dependent. DA is the best candidate for medical image segmentation by an advanced clustering technique. It is not sensitive to parameter tuning, and initialization problem, and is noise tolerant and guaranteed to converge.
Advanced clustering techniques can provide general solutions for effective segmentation of a broad range of medical images. All segmentation examples presented in section 6.3 use image intensity as the single feature to clustering algorithms to demonstrate the efficiency of the algorithms. In real applications, local property or connectivity of adjacent pixel can be embedded into segmentation to achieve more accurate segmentation [66, 67].
6.6 Acknowledgments
This work has been partially supported by funds from the Advanced Technology Program (ATP) (Grant No. 003644-0280-1999), Technology Development and Transfer Program (TDT) (Grant No. 003644-0217-2001) of the state of Texas, Kestrel Corporation, the NEI Grant No. 1 R43 EY14090-01 and the NSF Grant EIA9980296. We acknowledge Young I. Kim, M.D., and Mary Lucy M. Pereira, M.D., of Young H. Kwon’s (M.D., Ph.D.) team from University of Iowa Hospitals and Clinics for their contributions to manual segmentation of the stereo optic disk images. The authors are grateful to Daron Ferris, M.D., from the Medical College of Georgia for providing us with the cervigram images from the Guanacaste Project, Costa Rica, sponsored by the National Cancer Institute of USA.
Questions
1.What is the structure of adaptive fuzzy leader clustering (AFLC)?
2.Does AFLC have to initialize like k-means? If not, why?
Statistical and Adaptive Approaches for Optimal Segmentation |
307 |
3.How does AFLC dynamically adjust the number of clusters?
4.What is the difference between deterministic annealing (DA) and simulated annealing (SA)?
5.What is the DA cost function and what does it minimize?
6.What effect does the temperature reduction rate parameter have on DA clustering?
7.How does DA adjust the number of clusters?
8.What does mass-constrained DA mean?
9.What makes MS segmentation different from normal brain segmentation?
10.Judging from the examples given in the chapter, what are the performance differences among AFLC, DA, FCM, and k-means?
11.What is the limitation of clustering segmentation based on image intensity?
12.How is clustering in retinal optic disk/cup and blood vessel segmentation better than regular edge detection techniques?
13.Why is registration necessary in 3-D retinal disk/cup segmentation and how is it done?
14.How is the 3-D optic disk/cup map created?

308 |
Yang and Mitra |
Bibliography
[1]Pham, D. L., Xu, C. J., and Prince, L., A survey of current methods in medical image segmentation, Ann. Rev. Biomed. Eng., Jan 1998.
[2]Haralick, R. M. and Shapiro, L. G., Image segmentation techniques, Comput. Vision, Graphics Image Process., Vol. 29, No. 1, pp. 100–132, 1985.
[3]Fu, K. S. and Mui, J. K. A survey on image segmentation, Patt. Recogn., Vol. 13, pp. 3–16, 1981.
[4]Pal, N. R. and Pal, S. K., A review on image segmentation techniques, Patt. Recogn., Vol. 26, No. 9, pp. 1277–1294, 1993.
[5]Suri, J. S., Setarehdan, S. K., and Singh, S., eds., Advanced Algorithmic Approaches to Medical Image Segmentation, Springer-Verlag, London, 2002.
[6]Skarbek, W. and Koschan, A., Colour image segmentation: A survey, Technical Report 94-32, Technical University, Berlin, 1994.
[7]Lucchese, L. and Mitra, S. K., Color image segmentation: A state-of- the-art survey, Proc. Indian Nat. Sci. Acad. (INSA-A), Vol. 67, No. 2,
pp.207–221, 2001.
[8]Bezdek, J. C., Hall, L. O., and Clarke, L. P., Review of MR image segmentation techniques using pattern recognition, Med. Phys., Vol. 20,
pp.1033–1048, 1993.
[9]Clarke, L. P., Camacho, R. P., Velthuizen, M. A., Heine, J. J., Vaidyanathan, M., Hall, L. O., Thatcher, R. W., and Silbiger, M. L., Review of MRI segmentation: Methods and applications, Magn. Reso. Imaging, Vol. 13, No. 3, pp. 343–368, 1995.
[10]Zadeh, L. A., Fuzzy sets, Inf. Control Theory, Vol. 8, pp. 338–353, 1965.
[11]Zadeh, L. A., Outline of a new approach to the analysis of complex systems and decision processes, IEEE Trans. Syst., Man, Cybern., Vol. SMC-3, pp. 28–44, 1973.
Statistical and Adaptive Approaches for Optimal Segmentation |
309 |
[12]Jain, A. K., Murty, M. N., and Flynn, P. J. Data clustering: A review, ACM Comput Surveys, Vol. 31, No. 3, pp. 264–323, 1999.
[13]Rose, K. Deterministic annealing for clustering, compression, classification, regression, and related optimization problems, Proc. IEEE, Vol. 86, No. 11, 1998.
[14]Newton, S. C., Pemmaraju, S., and Mitra, S., Adaptive fuzzy leader clustering of complex data sets in pattern recognition, IEEE Trans. Neural Networks, Vol. 3, pp. 794–800, 1992.
[15]MacQueen, J., Some methods of classification and analysis of multivariate observations, In: Proceedings of 5th Berkeley Symposium on Math., Stat., and Prob., LeCam, L. M. and Neyman, J., eds., University of California Press, Berkeley, CA, pp. 281, 1967.
[16]Bezdek, J., Pattern Recognition with Fuzzy Objective Function Algorithms, Plenum Press, New York, 1981.
[17]Gonzalez, R. C. and Woods, R. E., Digital Image Processing, Addison Wesley, Reading, MA, 1992.
[18]Marroquin, J. L. and Girosi, F., Some extensions of the k-means algorithm for image segmentation and pattern recognition, Technical Report, MIT AI Lab., AI Memo 1390, Jan 1993.
[19]Fraley, C. and Raftery, A. E., How many clusters? Which clustering method? Answers via model-based cluster analysis, Technical Report No. 329, University of Washington, 1998.
[20]Ray, S., Turi, R. H., and Tischer, P. E., Clustering-based color image segmentation: An evaluation study, In: Proceedings of Digital Image Computing: Techniques and Applications, Brishbane, Qld., Austrlia, 6–8 Dec. 1995, pp. 86–92.
[21]Park, S. H., Yun, I. D., and Lee, S. U., Color image segmentation based on 3-D clustering: Morphological approach, Patt. Recogn., Vol. 31, No. 8, pp. 1061–1076, 1998.
[22]Weeks, A. R., and Hague, G. E., Color segmentation in the HIS color space using the k-means algorithm, In: Proceedings of the
310 |
Yang and Mitra |
SPIE—Nonlinear Image Processing VIII, San Jose, CA, Feb, 10–11, 1997,
pp.143–154.
[23]Wu, J., Yan, H., and Chalmers, A. N., Color image segmentation using fuzzy clustering and supervised learning, J. Electron. Imaging, pp. 397– 403, 1994.
[24]Pappas, T. N., An adaptive clustering algorithm for image segmentation, IEEE Trans. Signal Process., Vol. SP-40, pp. 901–914, 1992.
[25]Schmid, P., Segmentation of digitized dermatoscopic images by twodimensional color clustering, IEEE Trans. Med. Imaging, Vol. MI-18, No. 2, pp. 164–171, 1999.
[26]Carpenter, G. A. and Grossberg, S., A massively parallel architecture for a self-organizing neural pattern recognition machine, Comput Vision, Graphics Image Process., Vol. 37, pp. 54–115, 1987.
[27]Carpenter, G. and Grossberg, S. Art-2: Self organization of stable category recognition codes for analog input patterns, Appl. Opt., Vol. 26,
pp.4919–4930, 1987.
[28]Carpenter, G. and S. Grossberg, Art-3: Hierarchical search using chemical transmitters in self-organizing pattern recognition architectures, Neural Networks, Vol. 3, pp. 129–152, 1990.
[29]Grossberg, S., Embedding fields: A theory of learning with physiological implications, J. Math. Psychol., Vol. 6, pp. 209–239, 1969.
[30]Rumelhart D. E. and Hipser, D., Feature discovery by competitive learning, In: Parallel Distributed Processing, MIT Press, Cambridge, MA,
pp.151–193, 1986.
[31]Mitra, S. and Yang, S. Y., High fidelity adaptive vector quantization at very low bit rates for progressive transmission of radiographic images, J. Electron Imaging, Vol. 11, No. 4(Suppl. 2), pp. 24–30, 1998.
[32]Mitra, S., Castellanos, R., Yang, S. Y., and Pemmaraju, S., Adaptive clustering for image segmentation and vector quantization, In: SoftComputing for Image Processing, Editors: Pal, S. K., Ghosh, A., and Kundu, M. K., eds., Springer-Verlag, New York, 1999.
Statistical and Adaptive Approaches for Optimal Segmentation |
311 |
[33]Kirkpatrick, S., Gelatt, C. D., and Vecchi, M. P., Optimization by simulated annealing, Science, Vol. 220, pp. 671–680, 1983.
[34]Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H., and Teller, E., Equations of state calculations by fast computing machines,
J.Chem. Phys., Vol. 21, No. 6, pp. 1087–1091, 1953.
[35]Johnston, B., Atkins, M. S., Mackiewich, B., and Anderson, M., Segmentation of multiple sclerosis lesions in intensity corrected multispectral MRI, IEEE Trans. Med. Imaging, Vol. 15, pp. 154–169, 1996.
[36]Zijdenbos, A. P., Dawant, B. M., Margolin, R. A., and Palmer, A. C., Morphometric analysis of white matter lesions in MR images: Method and validation, IEEE Trans. Med. Imaging, Vol. 13, No. 4, pp. 716–724, 1994.
[37]Hall, L. O., Bensaid, A. M., Clarke, L. P., Velthuizen, R. P., Silbiger, M. S., and Bezdek, J. C., A comparison of neural network and fuzzy clustering techniques in segmenting magnetic resonance images of the brain, IEEE Trans. Neural Networks, Vol. 35, pp. 672–682, 1992.
[38]Clark, M. C., Hall, L. O., Goldgof, D. B., et al., MRI segmentation using fuzzy clustering-techniques, IEEE Eng. Med. Biol., Vol. 13, No. 5,
pp.730–742, 1994.
[39]Kamber, M., Collins, D. L., Shinghal, R., Francis, G. S., and Evans,
A.C., Model-based 3-D segmentation of multiple sclerosis lesions in dual-echo MRI data, Proc. SPIE Visual. Biomed. Comput., Vol. 1808,
pp.590–600, 1992.
[40]Jackson, E. F., Narayana, P. A., Wolinsky, J. S., and Doyle, T. J., Accuracy and reproducibility in volumetric analysis of multiple sclerosis lesions,
J.Comut. Assisted Tomog., Vol. 17, No. 2, pp. 200–205, 1993.
[41]Kapouleas, I., Automatic detection of white matter lesions in magnetic resonance brain images, Comput. Methods Programs Biomed., Vol. 32,
pp.17–35, 1990.
[42]Mitchell, J. R., Karlik, S. J., Lee, D. H., and Fenster, A., Automated detection and quantification of multiple sclerosis lesions in MR volumes of
