
Kluwer - Handbook of Biomedical Image Analysis Vol
.3.pdf370 |
Kybic and Unser |
9.4.8.5 Consequences of Finite Support
All what we said so far about expansion and reduction holds for infinite signals. To adapt the method for finite signals, we considered the following requirements: the expansion must be exact in the continuous sense, the projection identity must hold, reduction followed by expansion must conserve the length of the signal, and as much information as feasible should be conserved. These requirements are useful to guarantee the best possible use of the coarse-grid results at the fine-grid level and are absolutely indispensable for multigrid minimization.
Traditionally, one represents the signal with exactly one coefficient per sample and assumes that the signal outside the region of interest follows some known pattern, such as periodicity, or mirror-on-boundary conditions. We take the mirror-on-boundary condition as an example, but the same kind of problems appear for other boundary conditions, too. First, the signal is forced to be symmetric and thus flat at boundaries. Second, the boundary conditions for both expansion and reduction are only conserved for odd number of samples, otherwise the mirror position needs to change. Third, varying the length of the signal by one does not change the length of the reduced version which makes it impossible to recover the original length by expansion.
The centered pyramids [108] conserve the mirror position for even-length signals. Unfortunately, the expansion is no longer exact. Moreover, the constraint of the size of the image being a power of two, together with the integer step size h, seems to be too restrictive.
Because of these considerations, we decided to dissociate the number of B-spline coefficients from the length of the interest region. Initially, we extend the signal by (n − 1)/2 samples at each extremity which allows us to represent any spline of degree n without constraints. We never move the boundaries of our signal when expanding, although the number of B-splines might vary. In this way, expansion is always exact while it adds extra knots at each end. Reducing expanded signal recovers exactly the original. When reducing other signals, we need to extend them to be able to use our efficient filtering technique. For this, we choose to use the mirror-on-boundary conditions.
9.4.8.6 Image Size Change
The only trick when expanding and reducing the images is to adapt the deformation function accordingly. This is easily accomplished by multiplying the
Elastic Registration for Biomedical Applications |
371 |
coefficients by 2 when expanding and 0.5 when reducing. Thanks to our choice of the expansion and reduction operators, the origin of the image does not change.
9.4.8.7 Fast Spline Calculations
It is essential to take full advantage of the properties of splines. First, specialized routines are used to calculate the values of a B-spline of a specific order using a minimum number of operations. Second, as we are using tensor products of B-splines as our basis functions, many operations can be performed in a separable fashion, reducing the complexity of operations from O (kN ), where N is the number of dimensions and k the size of the data, to O (kN). This is the case for the prefiltering step required to find the B-spline coefficients, and also for the interpolation of values of a function given by its B-spline coefficients. Third, the compact support of B-splines simplifies many of the infinite sums in the expressions given earlier, reducing them to sums over just a small number of elements.
9.4.8.8 Stopping Criterion
To get a fast optimization algorithm, particular attention has to be paid to the stopping criterion. This holds for both GD and ML algorithms. Classically, the relative and absolute improvement of the criterion value is compared with a fixed threshold [59]. For our class of problems, it is often advantageous to base the stopping criterion on the changes c of parameter values. We stop when the step size falls below an a priori given threshold ε. The size of a step that fails gives an indication of the accuracy of the result and is therefore easy to set. Typically, we would use the threshold of ε = 10−1 10−3 pixels for the finest level and slightly more for coarser levels, as there is usually not enough details and coherence between levels.
9.4.8.9 Masking
A substantial gain in speed comes from considering only important pixels when calculating the data criterion (9.7) and its derivatives. It is possible to determine an a priori mask of significant pixels, for example, 10 50 % of the total

372 |
Kybic and Unser |
Figure 9.11: Example of a mask selecting 10 % of the pixels with the highest gradient values for the Lena image.
number of pixels, and to consider only those pixels in subsequent calculations. The contributions of individual pixels to the change of the criterion is directly proportional to the amplitude of the directional derivatives at the respective points, see (9.12). Therefore, a reasonable strategy is to construct the mask by thresholding the gradient of the image at each multiresolution level (see Fig. 9.11).
9.4.9 Invertibility and Regularization
In some applications, it is useful to add an extra regularization term Er to the difference measure E, and to look for a minimum of the combined criterion
Ec = E + γ Er |
(9.21) |
The factor Er is used to make the solution well-posed, or to privilege likely solutions based on our a priori knowledge.
First, we consider a penalty term designed to enforce the invertibility of the deformation, generalizing the concept from [95] and similar to [113]. Its moti-
vation comes from the fact that if the Jacobian det( g) is positive everywhere,
x
Elastic Registration for Biomedical Applications |
373 |
then the deformation g is locally invertible. Evaluating this constraint at pixel coordinates and converting the strict constraints into soft ones using a barrier function yields the following penalty term
Ep = |
e− |
α det( xg(i)) |
(9.22) |
|
|||
i I |
|
|
|
|
|
|
|
Experience shows that, for typical data, this term is never important at the solution point (to which the optimization converges). It mostly becomes useful at the beginning of the optimization process when the trial points vary a lot, especially with some optimizers. In such cases, the penalty term forces the algorithm to stay in the region of invertible deformations.
Depending on the particular task and the expected properties of the solution, various regularization terms can be used. We investigated, for example,
a stabilizer penalizing non-linear deformations
|
|
|
|
∂2 gk |
|
2 |
|
Et = |
N |
|
|
dx |
(9.23) |
||
k,l,m |
1 |
∂ xl ∂ xm |
|||||
|
= |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
and a very simple norm measuring the distance of g from identity through the coefficients c
Ed = !cj!2 (9.24)
j
When the corresponding weight γ is small, the regularization mainly smoothes the deformation function in places where little information is present in the images. As it gets bigger, the regularization gradually overrides the data term and the deformation tends towards a smooth function in the sense of the particular regularization. An alternative to regularization is the virtual spring mechanism described in section 9.4.7.
9.4.10 Experiments
We now illustrate the application of the presented algorithm to various problems involving medical images of several modalities. We refer the interested reader to [88,95,96], where we study in detail the accuracy, speed and robustness of the algorithm by means of a comprehensive series of experiments in a controlled environment.

374 |
Kybic and Unser |
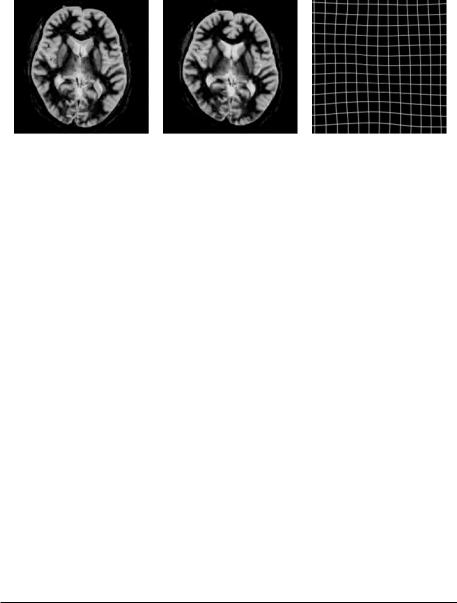
Figure 9.12: The original slice of anatomical MRI brain image ((top left), original superimposed over the true deformation (top right), the recovered deformation versus the true deformation (bottom left), and the mask used to calculate the warping index (bottom right). (Color Slide)
9.4.10.1 Registration of Artificially Deformed MRI Brain Slices
To illustrate the behavior of the algorithm, we show its performance when recovering a known deformation of a 2D slice of an anatomical spin-echo MRI volume of the brain.9 We use here artificially-deformed images because the knowledge of the ground truth permits us to better judge the performance of the algorithm.
The original image of size 256 × 256 pixels is shown in Fig. 9.12, top-left. We use a cubic spline control grid with one knot for every 32 pixels. We warp the image with a deformation belonging to the warp space and consisting of displacements up to 15 pixels.10 The warped image is superimposed on the original in Fig. 9.12, top-right. Then the automatic registration algorithm is run with the stopping threshold set to 0.5 pixels for all levels except the last, where we set it to 0.1 pixels. The recovered deformation was used to warp again the original image. Its warped version is shown superimposed on the image warped
9 First author’s brain. Images courtesy of Arto Nirkko from Inselspital Hospital, Bern,
Switzerland.
10 Approximately 14 mm.

Elastic Registration for Biomedical Applications |
375 |
|
900 |
|
|
|
|
criterion |
|
|
|
|
|
|
|
|
|
|
800 |
|
|
|
|
|
|
|
700 |
|
|
|
|
|
|
|
600 |
|
|
|
|
|
|
SSD |
500 |
|
|
|
|
|
|
mean |
400 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
300 |
|
|
|
|
|
|
|
200 |
|
|
|
|
|
|
|
100 |
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
0 |
10 |
20 |
30 |
40 |
50 |
60 |
|
|
|
|
iteration number |
|
|
|
|
5 |
|
|
|
warping index |
|
|
|
|
|
|
|
|
||
|
4.5 |
|
|
|
|
|
|
|
4 |
|
|
|
|
|
|
|
3.5 |
|
|
|
|
|
|
|
3 |
|
|
|
|
|
|
pixels |
2.5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
1.5 |
|
|
|
|
|
|
|
1 |
|
|
|
|
|
|
|
0.5 |
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
0 |
10 |
20 |
30 |
40 |
50 |
60 |
|
|
|
|
iteration number |
|
|
|
|
900 |
|
|
|
|
criterion |
|
|
|
|
|
|
|
|
|
|
800 |
|
|
|
|
|
|
|
700 |
|
|
|
|
|
|
|
600 |
|
|
|
|
|
|
SSD |
500 |
|
|
|
|
|
|
mean |
400 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
300 |
|
|
|
|
|
|
|
200 |
|
|
|
|
|
|
|
100 |
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
0 |
5 |
10 |
15 |
20 |
25 |
30 |
|
|
|
|
time in seconds |
|
|
|
|
5 |
|
|
|
warping index |
|
|
|
|
|
|
|
|
||
|
4.5 |
|
|
|
|
|
|
|
4 |
|
|
|
|
|
|
|
3.5 |
|
|
|
|
|
|
|
3 |
|
|
|
|
|
|
pixels |
2.5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
1.5 |
|
|
|
|
|
|
|
1 |
|
|
|
|
|
|
|
0.5 |
|
|
|
|
|
|
|
0 |
|
|
|
|
|
|
|
0 |
5 |
10 |
15 |
20 |
25 |
30 |
|
|
|
|
time in seconds |
|
|
|
Figure 9.13: The evolution of the optimization process. The left column displays the evolution with respect to the number of iterations, while the right column represents the same quantity respect to time. The first row shows the SSD criterion E, the second row the warping index . The steep (step) changes correspond to the changes in the model and image resolutions. We observe good correlation between all four graphs.
with the true deformation in Fig. 9.12, bottom-right. We note that the deformation was well recovered with no perceptible difference.
The spatial distribution of the resulting geometrical error is shown in Fig. 9.14. The maximum error is about 1.5 pixels, while the mean geometric error (warping index [36]) over the total of the brain is about 0.4 pixels. We generally observe that the error is concentrated in areas with little detail in the image. Other high-contrast regions, such as edges, are resolved much more precisely than indicated by the value of , often with subpixel accuracy. On the other hand, the agreement in the zones with low-contrast is worse and often only coincidental, since there is little or no information to guide the algorithm.
The evolution of the optimization can be studied from the graphs in Fig. 9.13. We observe the steady and correlated descent of the observable criterion being

376 |
Kybic and Unser |
Figure 9.14: The geometrical error after registration (green) with superposed contours of the original MRI image (red). The maximum (green) intensity corresponds to an error of 1.5 pixels. (Color slide.)
optimized (E) and of the warping index ( ), the quantity measuring the quality of the registration. The abrupt changes in the curves are caused by the transitions between levels of the multiresolution progression; they are small thanks to the
accuracy of the spline model. |
|
Note that the final values of both E and |
depend strongly on the pre- |
set stopping threshold, which in turn influences the optimization time. The threshold value is a subjective compromise between the accuracy and computation time. It is perfectly possible to stop optimizing only after 7 s and
skip the finest resolution level altogether, if the precision of = 0.7 pix-
els is acceptable. On the other hand, after about 4 more minutes of iteration, the error descends to less than 10−4 pixels. However, in the authors’
opinion, such super subpixel accuracy is almost never achievable on real images, because of the noise and the unknown characteristics of the acquisition process.
9.4.10.2 Registration of True Medical Data
Finally, we give a representative list of medical imaging registration tasks where the described algorithm was successfully used:
Registration of ECD11 and Xenon inhalation SPECT images [114] in the view of atlas creation [115].
11 ECD (Technetium Ethylene Cysteine Diethylester) is a radioactively marked intra-
venously injected agent.
Elastic Registration for Biomedical Applications |
377 |
(a) |
(b) |
(c) |
Figure 9.15: The superposition of the slices of anatomical MRI images before the registration (a), after the registration (b), and the resulting deformation field
(c). Quadratic splines were used with knot spacing of h = 64. (Color slide.)
Intersubject registration of anatomical (spin-echo) MRI images,12 Fig. 9.15.
Registration of MRI images from a heart beat sequence,13 see Fig. 9.16. The extracted deformation field can be used to extract trajectories of various points in the heart and to further determine the velocity and derived parameters, such as the accumulated displacement and strain, which is important for diagnostic purposes.
Registration of standard 2D ultrasound sequences of the heart [116].
Motion compensation for a sequence of myocardial perfusion MRI images14 [117, 118], enabling the time profiles of the intensities at each tissue point to be reliable calculated (Fig. 9.17). Virtual springs were used in this case.
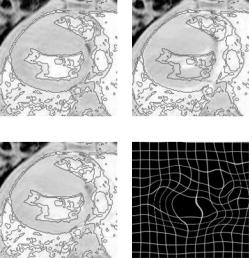
Registration of two 3D computer tomography (CT) head volumes,15 see Figs. 9.18 and 9.19. We worked on a reduced volume data (128 × 128 × 45
instead of the original 512 × 512 × 45) with control knots placed every
8 × 8 × 8 voxels and the process took about 10 minutes to complete.16
12Images courtesy of Arto Nirkko, Inselspital Hospital, Bern, Switzerland.
13LECB, NIH, http://www-lecb.ncifcrf.gov/flicker/
14Courtesy of J.-P. Vallee,´ Unite´ d’imagerie numerique,´ University Hospital, Geneva,
Switzerland.
15 Images courtesy of Philippe Thvenaz, EPFL, Lausanne, Switzerland. The images were acquired using the same machine and the same protocol, but not preregistered.
16 On a 700 MHz Pentium based computer. Registering directly the undecimated volumes on the same computer takes about three hours with very minor increase in quality as relatively smooth deformations are sought. We are currently working on a optimized reimplementation of the algorithm that should reduce these times considerably.
378 |
Kybic and Unser |
(a) |
(b) |
(c) |
(d) |
Figure 9.16: The reference MRI image from a heart sequence with superimposed contours (a). The same contours over another image (the test image) from the same sequence before the registration (b) and after (c). The deformation field (d). Quadratic splines were used with knot spacing of h = 64, image size was 256 × 256 pixels. (Color slide.)
9.4.11 Summary of Elastic Registration
The algorithm that has been described is a state-of-the-art example of what is available in the field of fully automatic elastic pixel-based registration. It contains many features that have been proposed in the literature and it has been streamlined for an efficient execution.
Elastic registration has numerous applications in the biomedical imaging field, all based on the basic notion of aligning two images with one other, be it intersubject, intrasubject, or intermodality. It can be used for motion and deformation detection. The deformation field itself can be used for deformation and motion compensation as well as for quantitative measurements.
The criterion, the deformation model, the regularization (penalty), and the optimization algorithm should be all adapted to the task at hand for optimum results.
By producing a specialized program taking advantage of a specific configuration, the runtime can be probably decreased by an additional factor of 10.

Elastic Registration for Biomedical Applications |
|
379 |
|
6 |
9 |
11 |
14 |
uncorrected (adj.) original |
|
|
|
corrected (adj.) |
|
|
|
uncorrected (first) |
|
|
|
corrected (first) |
|
|
|
Figure 9.17: The first line presents original images number 6,9,11, and 14 from a sequence of originally 60 images of myocardical perfusion MRI. The second line presents the difference images between the original images and their immediate predecessors; movement artifacts can be clearly seen. On the third line you can see the difference images from the motion corrected sequence using our algorithm; the movement artifacts are significantly reduced. The same effect is also visible comparing the differences of the sequence images with the first image of the sequence on the original (fourth line) and corrected (fifth line) sequences.
This, together with the constant advances in computer technology will enable truly interactive operation of automatic and semi-automatic elastic image registration with numerous applications in medicine, biology, and any other field where deformed images need to be compared.
