
John Wiley & Sons - 2004 - Analysis of Genes and Genomes
.pdf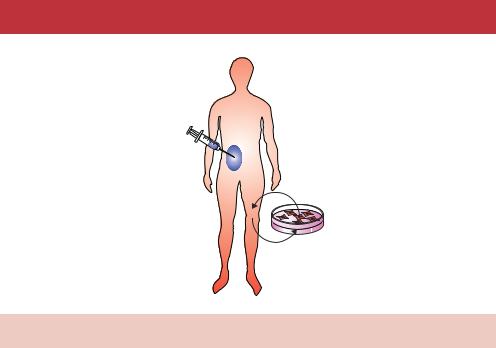
398 ENGINEERING ANIMALS 31
In vivo
Ex vivo
Figure 13.7. In vivo and ex vivo human gene therapy. See the text for details
cultured in vitro for extended periods. Therefore, disorders of the blood system may be treated ex vivo through the isolation and culturing of bone marrow stem cells. These cells can be modified in vitro and the resulting transgenic cells can then be returned to the patient. The modified stem cells will then produce the various modified differentiated cells that may cure the disease. In vitro manipulation of the cells permits the use of a wide variety of methods to insert the transgene – e.g. viral infection, injection and other methods (see Chapter 12). Other cells and tissues are less amenable to ex vivo treatment. For example, lung epithelial cells – whose function is severely impaired in cystic fibrosis patients – grow very poorly in culture. Even if they could be cultured, it would not be possible to repopulate an affected lung with transgenic lung epithelial cells. Therefore, diseases such as cystic fibrosis must be treated in vivo so that the cells of the defective lung can be modified. This limits the type of transgene insertion that can take place. Viruses, e.g. adenovirus that naturally infect epithelial cells, are usually used to transport the transgene into the affected cells.
13.5Examples and Potential of Gene Therapy
The history of human gene therapy trails is not a particularly happy one. With one notable exception (see below), the effect of introducing a gene into cells rarely promotes more than a transient relief from the symptoms of the disease being treated. Worse still, there have been highly publicized cases where gene
13.5 EXAMPLES AND POTENTIAL OF GENE THERAPY |
399 |
|
|
therapy trial patients have suffered as a consequence of the treatment itself. For example, in 1999 an 18-year-old gene therapy trial volunteer from Philadelphia, Jesse Gelsinger, died following a gene therapy trial (Teichler Zallen, 2000). Gelsinger had an ornithine transcarbamylase (OTC) disorder, a rare genetic defect of the liver that renders the body unable to clear ammonia from the bloodstream. He was treated with an adenoviral vector as a mechanism to insert a healthy copy of the gene into his liver, but the virus itself elicited a massive immune reaction that resulted in his death. Cases such as this graphically illustrate the need for the development of vectors characterized by maximum transfection efficiency and minimal toxicity.
Some gene therapy successes have, however, been noted. Children born with severe combined immune deficiency, X-SCID, have a poor prognosis using traditional medicines. The disease is caused by a mutation on the X chromosome in the gene encoding the gamma chain (γ c) of the interleukin-2 receptor. Mutations in this gene prevent two types of white blood cell, the T-cells and natural killer cells, from developing normally (Sugamura et al., 1996). With little or no defence against infection, sufferers usually die within the first year of life unless a bone marrow donor can be found. Stem cells were collected from the bone marrow of an affected infant and treated with a retrovirus carrying a wild-type copy of the γ c gene (Cavazzana-Calvo et al., 2000). When the transgenic stem cells were returned to the infant they were capable of generating all of the cells required for a fully functional immune system for at least 10 months (Fischer, Hacein-Bey and Cavazzana-Calvo, 2002). Removing the bone marrow cells from the body prior to infection with the retrovirus eliminates the danger of acute reaction to the virus itself, and also ensures that the virus only infects the correct cells. Repopulating the immune system with a relatively small number of transgenic bone marrow cells may also cause problems. The treatment specifically selects for proliferating cells and may therefore increase the risk of bone marrow related cancers. It has been noted that some of patients treated in this way develop leukaemia (one out of 10 patients successfully treated), attributed to a result of the integration of the foreign DNA fragments into the genome at random locations. In this case, the retrovirus inserted the therapeutic gene into the regulatory region of a gene called Lmo2 on chromosome 11 (Gansbacher¨ et al., 2003). The activation of the therapeutic gene appeared to cause the expression of Lmo2 which is an oncogene (Davenport, Neale and Goorha, 2000). Even with these problems, these experiments represent the only example to date where a patient is apparently completely cured using gene therapy.
Some of the problems associated with random integration of the transgene during gene therapy may be addressed by utilizing site-specific recombination

400 ENGINEERING ANIMALS 31
systems. For example, DNA fragments have been constructed such that they contain a therapeutic gene adjacent to the recognition sequence of a site-specific recombinase enzyme. If these DNA fragments are injected into the tail veins of mice together with a DNA fragment encoding the integrase, then site-specific genomic integration of the transgene occurs (Olivares et al., 2002). This could be developed from the mouse model into a human therapy.
Other gene therapy trails are currently ongoing for both genetic and nonheritable diseases.
• Haemophilia B. Sufferers lack the gene for factor IX, a critical agent in the blood clotting process. Parvoviruses have been used to insert the missing gene into skeletal muscle cells (High, 2001). The cells then generate the missing factor, thereby removing the need for daily injections of the protein itself.
•Cancer. Some cancer treatments may be amenable to gene therapy (Wadhwa et al., 2002). Modified viral vectors can be used to prime the immune system to attack cancer cells, while other approaches employ viruses to carry suicide genes into the cancer cells.
•HIV. Specifically engineered HIV may eventually be recruited to help control HIV-1 infection (Statham and Morgan, 1999).
Currently, the promise of gene therapy remains just that. Even single gene defect diseases can manifest themselves as deficiencies in a wide variety of different cell types. Being able to correct the defect in one cell type may not be sufficient to cure the disease fully. However, the development and refinement of transgene delivery systems, combined with advances in our understanding of stem cells may generate many more opportunities in the future where gene therapy may be clinically important.


402 GLOSSARY
Centromere – the point or region on a chromosome to which the spindle attaches during mitosis and meiosis
Chromatid – one of the usually paired and parallel strands of a duplicated chromosome joined by a single centromere
Chromatin – a complex of DNA and proteins in the nucleus of a cell
Chromatin immunoprecipitation (ChIP) – a method for identifying proteins bound to particular sequences of DNA
Chromosome – a discrete unit of the genome that is visible as a morphological entity during cell division. Each chromosome is a single DNA molecule
Chromosome walking – the sequential isolation of clones carrying overlapping DNA sequences that allows the sequencing of large regions of the chromosome from a single starting point
Clone – an organism, cell or molecule produced from a single ancestor Cloning vector – a plasmid or phage that is used to carry inserted foreign DNA
Codon – the triplet of nucleotides that result in the insertion of an amino acid or a termination signal into a polypeptide
Codon usage – the frequency at which amino acid codons are used for the production of proteins
Complementary – the sequences on one strand of a nucleic acid molecule can bind to their complementary partners on another strand. A = T, G = C
Conjugation – the transfer of all or part of a chromosome that occurs during bacterial mating
Conservative replication – a disproved model for DNA synthesis in which the newly synthesized DNA strands bind to each other
Contig – a continuous sequence of DNA produced from a number of smaller, overlapping fragments
Cosmid – a plasmid onto which phage lambda cos sites have been inserted. Consequently, the plasmid DNA can be packaged in vitro into the lambda phage coat Cytological map – a type of chromosome map where genes are located on the basis of
the effect that chromosome mutations have on staining patterns
Cytosine – a pyrimidine base found in DNA and RNA. Cytosine bases pairs with guanine
Denatured – in DNA, the conversion of the double-stranded form to a single-stranded form. In proteins, the conversion from an active to an inactive form
Differential display – a technique to visualize difference in the expression of genes from different sources
Dinucleotide – the joining of two nucleotides through the formation of a phosphodiester linkage
Dispersive replication – a disproved model of DNA synthesis in which a random interspersion of parental and new segments are found in daughter DNA molecules
DNA – deoxyribonucleic acid
DNA ligase – the enzyme that catalyses the formation of a phosphodiester bond between two DNA chains
DNA polymerase – the enzyme that synthesizes new DNA strands from a DNA template DNA topoisomerase – an enzyme that changes the linking number of DNA molecules

GLOSSARY 403
Electrophoresis – the application of an electric current to separate molecules (as proteins and nucleic acids) through a gel
Electroporation – a physical way to introducing DNA into cells using an electric current Epitope – the molecular region on the surface of an antigen capable of eliciting an
immune response
Euchromatic DNA – the gene-rich areas (including both exons and introns) of a genome Exon – a segment of a gene that is represented in the mature mRNA
Expressed sequence tag (EST) – small pieces of cDNA sequence generated by sequencing either one or both ends of an expressed gene
Expression vector – a cloning vector designed so that the genes cloned into it may be transcribed and translated
F episome – a large extrachromosomal circular double-stranded DNA molecule that carries bacterial fertility genes
FISH – fluorescent in situ hybridization
Frame-shift mutation – occurs when the coding sequence of a gene contains a deletion or insertion of bases that are not in multiples of three. This changes the reading frame in which translation occurs
Functional complementation – the identification of genes from one organism by their ability to counteract the defect caused by the lack of a gene in another organism
Gene – a discrete unit of genetic information that is required for the production of a polypeptide. It includes the coding sequence, the promoter and terminator, and introns
Gene knockout – the removal of a gene from the genome
Gene knock-down – the use of silencing techniques to reduce or eliminate the expression of a particular gene
Genetic engineering – the deliberate modification of the characters of an organism by the manipulation of DNA and the transformation of certain genes
Genetic marker – any DNA sequence that can be used to identify a gene or phenotypic trait associated with it
Genetic switch – the control of transcription in response to particular signals Genome – the genetic make-up of an organism
Genomic library – a collection of DNA fragments, derived from the genome of an organism, cloned into a vector
Guanine – a purine base found in DNA and RNA. Guanine base pairs with cytosine HAC – human artificial chromosome
Helper phage – provides certain functions, that are absent from a defective phage, to allow complete phage replication during a mixed infection
Hemimethylated DNA – DNA that contains methylated bases on one strand only Heterochromatic DNA – regions of the chromosome that are highly condensed and not
transcribed
Heterozygous – an individual with different alleles at a particular genomic locus Histone – a DNA binding protein that forms part of the nucleosome
Holliday junction – a central intermediate formed during recombination
Homologous recombination – DNA crossovers that occur between two homologous DNA molecules

404 GLOSSARY
Homology – a similarity often attributable to common origin
Homozygous – an individual with the same allele at a particular genomic locus Hybridization – the pairing of complementary nucleic acids (DNA – DNA or DNA –
RNA)
Hyperchromic effect – the increase in optical density that occurs when DNA become single stranded
Insertional inactivation – the destruction of the function of a gene by cloning a DNA sequence into it
Intron – a segment of DNA that is transcribed, but is removed from the transcript during splicing
Isopycnic centrifugation – the separation of the components of a mixture on the basis of differences in density
Isoschizomer – different restriction enzymes that recognize the same target DNA sequence
Karyotype – the chromosome content of an organism
Kozak sequence – the consensus sequence for the initiation of translation. A binding site for the small subunit of the ribosome
lac operon – the genes required in bacteria for the metabolism of lactose
Lagging strand – short DNA fragments are replicated discontinuously in a 5 – 3 direction and later covalently joined
Lariat – an intermediate of RNA splicing
Leading strand – the strand of DNA that is synthesized continuously during replication in a 5 – 3 direction
Melting temperature (Tm) – the temperature at the mid-point of DNA denaturation Methylated DNA – the modification of DNA bases by the addition of methyl (CH3)
groups
Microarray – sets of miniaturized reaction areas that may also be used to test the binding of DNA fragments
Microsatellites – highly polymorphic DNA markers comprised of a variable number of tandem repeats
Mini-prep – the purification of extra-chromosomal DNA (usually a plasmid) from a small culture (e.g. 1.5 mL) of bacterial cells
Monocistronic – an RNA molecule that codes for one protein
Monoclonal antibody – a highly specific protein that can bind to a single epitope within an antigen
Morula – a globular solid mass of cells formed by the cleavage of a zygote that precedes the blastula
mRNA – messenger RNA
Northern blot – a technique for the separation of RNA molecules through agarose gels followed by detection of specific RNAs through hybridization with singlestranded DNA
Nucleoside – a purine or pyrimidine base combined with either deoxyribose or ribose found in DNA or RNA
Nucleosome – the basic structural unit of chromatin
GLOSSARY |
405 |
|
|
Nucleotide – a purine or pyrimidine base combined with either deoxyribose or ribose and a phosphate group found in DNA or RNA
Oligonucleotide – a short molecule of single-stranded DNA, usually synthesized chemically
Oocyte – an egg before maturation
Opine – a product produced by the condensation product of an amino acid with either a keto-acid or a sugar
Origin of replication – the nucleotide sequence at which DNA synthesis (replication) is initiated
PAC – P1 derived artificial chromosome. A vector, based on the bacteriophage P1 genome, used to clone large DNA fragments in E. coli
Palindromic – a sequence of DNA that read on one strand in a 5 – 3 direction is the same as than on the other strand in a 5 – 3 direction
PCR – polymerase chain reaction – cycles of DNA denaturation, primer annealing and extension with DNA polymerase lead to a amplification of the target DNA sequence
PFGE – Pulsed field gel electrophoresis Phage – bacteriophage – a bacterial virus
Phage display – a technique that fuses peptides to capsid proteins on the surface of phages. Libraries of phage displayed peptides may be screened for binding to specific ligands
Phenotype – the functional and structural characteristics of an organism as determined by the interaction of the genotype with the environment
Pilus – a filament-like projection from the surface of a bacterial cell
Phosphodiester bond – a covalent bond between the 5 phosphate group on one nucleotide and the 3 hydroxyl group on an adjacent nucleotide
Plasmid – an autonomous self-replicating extra-chromosomal closed-circular DNA Polycistronic – an mRNA molecule that contains more than one coding region Polyclonal antibody – a mixture of many antibodies, each raised against the same
antigen
Polynucleotide – long chains of nucleotides linked together by phosphodiester bonds
Polypeptide – a chain of amino acids connected by peptide linkages
Polysome – multiple ribosomes actively translating a single mRNA molecule into polypeptides
Primer – a short DNA or RNA sequence that is paired with one strand of DNA and provides a free 3 hydroxyl group at which DNA replication can initiate
Promoter – a DNA sequence which serves as the binding site for transcription factors and RNA polymerase during the initiation of transcription
Pronucleus – the haploid nucleus of an egg or sperm cell prior to fertilization, and immediately after fertilization before the sperm and egg nuclei have fused into a single diploid nucleus
Propeller twist – the rotation of individual DNA base pairs within the double helix Purine – a nitrogen containing, double-ring basic compound that occurs in nucleic
acids. The purines in DNA and RNA are adenine and guanine

406 GLOSSARY
Pyrimidine – a nitrogen containing, single-ring basic compound that occurs in nucleic acids. The pyrimidines in DNA are cytosine and thymine, and those in RNA are cytosine and uracil
Quiescence – a dormant phase of cell growth. All but the most basic functions of a cell or group of cells have stopped, usually in response to an unfavourable environment. The cell remains dormant until its surroundings are more favourable
Replicon – part of the genome in which replication is initiated. Contains origins of replication
Restriction enzyme – (or restriction endonuclease) recognizes short, often palindromic, DNA sequences (recognition sites) and cleaves the DNA
Restriction map – a linear map of the restriction enzyme recognition sites with a DNA molecule
Reverse transcriptase – the enzyme that synthesizes DNA from RNA templates Reverse transcription – the synthesis of DNA from RNA
RFLP – restriction fragment length polymorphism – changes in DNA sequence that result in altered lengths of DNA when cleaved with a restriction enzyme
Ribosome – RNA-rich cytoplasmic granules that are sites of protein synthesis RNA – ribonucleic acid
RNA interference (RNAi) – the process in which the introduction of double-stranded RNA into a cell inhibits the expression of genes
RNA polymerase – the enzyme that synthesizes RNA from a DNA template
RT-PCR – a method for the amplification of a specific mRNA. Reverse transcriptase is used to form a cDNA which is then amplified using PCR
Semi–conservative replication – the separation of the strands of DNA during replication with each acting as a template for the synthesis of a new complementary DNA strand Shine-Dalgarno sequence – an mRNA sequence that precedes the translation initiation
codon and is complementary to a ribosomal RNA
Signal sequence – a short amino acid sequence that determines the localization of a protein within the cell
Silent mutation – a mutation with DNA that does not cause an alternation to the encoded amino acid sequence
Single-nucleotide polymorphisms (SNPs) – single-base-pair variations scattered within the genetic code of the individuals within a population
siRNA – small interfering RNAs. Synthetic short double-stranded RNA molecules that inhibit gene expression
Site-specific recombination – occurs between two specific, but not necessarily homologous, DNA sequences
Southern blot – a technique for the separation of DNA molecules through agarose gels followed by detection of specific DNAs after denaturation through hybridization with single-stranded DNA
Spliceosome – a complex consisting of RNA and small nuclear ribonucleoproteins (snRNPs). The spliceosome splices RNA transcripts by excising introns and ligating the ends of exons
Splicing – the removal of introns and joining of exons to form a mature mRNA

GLOSSARY 407
Stem cell – a cell from the embryo, foetus or adult that has capability to reproduce itself. It can give rise to specialized cells that make up the tissues and organs of the body
Supercoiling – the way in which closed-circular DNA crosses over its own axis in three-dimensional space
T-DNA – the portion of the Agrobacterium tumefaciens Ti plasmid that is inserted into the genome of the host plant cell
TA cloning – a method for cloning PCR products that relies on the addition of template independent A residues to PCR products by Taq DNA polymerase
Telomere – the repetitive DNA sequences at the end of a chromosome
Terminal transferase – an enzyme that catalyses the addition of nucleotides to the 3 -terminus of DNA
Terminator – the sequence of DNA that causes RNA polymerase to stop transcription Terminator technology – plants are engineered so when crops are harvested, all new
seeds produced are sterile
Thymine – a pyrimidine base found in DNA. Thymine base pairs with adenine Transcription – the synthesis of RNA from a DNA template
Transfection – the addition of DNA to eukaryotic cells
Transformation – the addition of DNA to bacteria. In eukaryotes, this term also refers to the state of cells that undergo tumour-like growth
Transforming principle – the factor identified by Griffith as being able to convert one bacterial type to another
Transgenic animals – created by inserting DNA sequences into the germ line through addition to the egg
Transition mutation – a mutation in DNA in which one purine – pyrimidine base pair is changed to a different purine – pyrimidine base pair
Translation – the synthesis of protein using an mRNA template
Transversion mutation – a mutation in DNA in which a purine – pyrimidine base pair is changed to a pyrimidine – purine base pair
Trinucleotide – the joining of three nucleotides through phosphodiester linkages Two-hybrid screen – a technique to identify proteins that are able to interact with
each other
Uracil – a pyrimidine base usually only found in RNA. Uracil bases pairs with adenine Variable number tandem repeats (VNTRs) – different numbers of tandemly repeated
DNA sequences at a given locus.
Vector – a DNA molecule that possesses the ability to self-replicate. Used to introduce foreign DNA into host cells, where it is replicated autonomously in large quantities Western blot – the detection of specific proteins following electrophoresis using anti-
bodies
Xenobiotic – a compound with a chemical structure that is foreign to an organism Xenotransplantation – the surgical transplantation of tissue or organs from an individ-
ual of one species into an individual of another species YAC – yeast artificial chromosome
Zygote – a cell formed by the union of two gametes (egg and sperm)
