
Micro-Nano Technology for Genomics and Proteomics BioMEMs - Ozkan
.pdf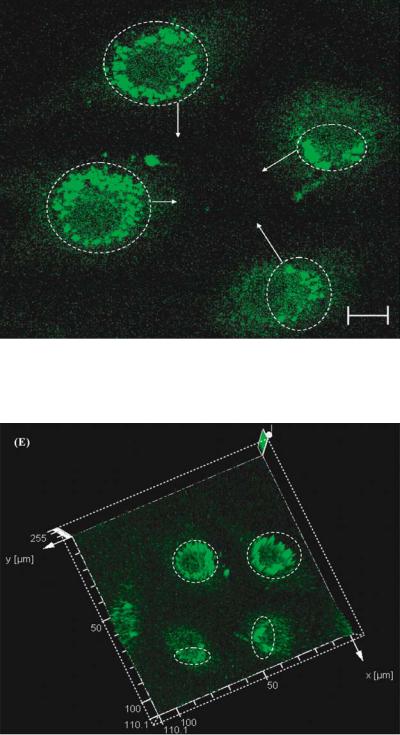
(D)
Localization of free potassium ions
5 m
FIGURE 3.15(D). Physiological effect of pyrethroid at 890 ppb on osteoblasts. The cells are stained with PBFI stain for visualizing free potassium ion transients. The confocal optical micrograph that shows the presence of potassium bundles after exposure to PBFI stain. This behavior correlates to the in-vitro behavior of large cell populations after exposure to organophosphates.
FIGURE 3.15(E). Three dimensional vector representation of the fluorescence intensity. The circled areas represent the high intensity peaks. The high intensity peaks are distributed uniformly near the cell membrane. These areas correspond to the areas of localization of the free potassium ions released due to the exposure to pyrethroid.

MICROARRAY AND FLUIDIC CHIP FOR EXTRACELLULAR SENSING |
93 |
FIGURE 3.15(F). Single osteoblasts stained with Hoechst 33382 stain after being exposed to pyrethroid at 890 ppb concentration for a period of one sensing cycle. The circled areas indicate the regions where the nucleic matter has condensed due to apoptosis. This behavior correlates to the in-vivo apoptotic behavior in osteoblasts after exposure to organophosphates at physiologically relevant concentrations.
the mechanical behavior of the bone. The bone samples became less brittle as the time of exposure to EDTA increased [8]. This translates into intracellular calcium emission after exposure to EDTA. His can be imaged by using fluo-3. This is a calcium indicator that binds to the generated free calcium ions. Figure 3.16(d) is the confocal optical micrograph that shows the calcium transients due to cell exposure to EDTA at 280 ppb for a period of one sensing cycle. Figure 3.16(e) is the three dimensional vector transformation of the confocal optical micrograph. The z axis gives the intensity levels. The fluorescence intensity peaks correspond to the locations of accumulation of calcium ions. EDTA also results in the DNA ladder formation. This occurs due to the clumping of the nucleic matter that happens due to the on-set of apoptosis [135]. Nucleic acid fragmentation is observed in the patterned osteoblasts exposed to 280 ppm concentration EDTA and stained with Hoechst 33382. Figure 3.16(f) is the confocal optical micrograph indicating this behavior.
3.35. DISCUSSION AND CONCLUSIONS
The past decade has seen great advancements in the field of bioanalysis along many fronts. Among the most rapidly advancing of these fronts is the area of biosensing, whether it is single analyte detection methods or multiarray-based biochip technology. The 1990s have seen the development of biosensors for many different analyses, and even seen them begin to advance to clinical and in some cases commercially available technologies [78, 97].
This great interest in the field of biosensors and biochips has revealed a great deal of information about the biology of all living things and may eventually provide an easy method
94 |
MIHRIMAH OZKAN ET AL. |
(A)
Intracellular calcium transients localized near the nucleus
5 m
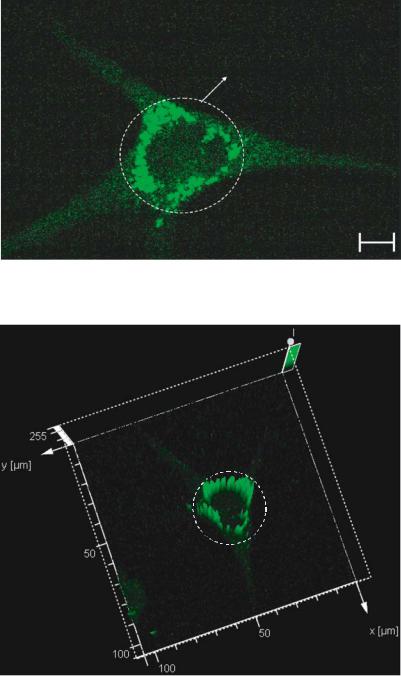
FIGURE 3.16(A). Physiological effect of EDTA at 180 ppm on individual neurons. The cells are stained with fluo-3 for visualizing free calcium ion transients. The calcium transients are localized in areas near the nucleus.
(B)
FIGURE 3.16(B). Three dimensional vector representation of the fluorescence intensity. The circled area represents the high intensity peaks. The high intensity peaks are distributed uniformly near the nucleus. These areas correspond to the areas of localization of the free calcium ions released due to the exposure to EDTA.
MICROARRAY AND FLUIDIC CHIP FOR EXTRACELLULAR SENSING |
95 |
(C)
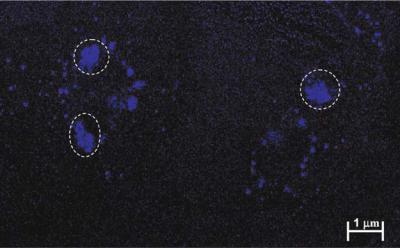
FIGURE 3.16(C). Single neurons stained with Hoechst 33382 stain after being exposed to EDTA at 180 ppm concentration for a period of one sensing cycle. The circled areas indicate the regions where the chromatin has condensed due to apoptosis.
for people to one day test themselves for certain illnesses at home or aid in the understanding of genetically transmitted illnesses. For practical medical diagnostic applications, there is currently a strong need for a truly integrated biochip system that comprises probes, samplers, detector as well as amplifier and logic circuitry. Such a system will be useful in physician’s offices and could be used by relatively unskilled personnel.
Biochip technologies offer a unique combination of performance capabilities and analytical features of merit not available in any other bioanalytical system currently available. With its multichannel capability, biochip technology allows simultaneous detection of multiple biotargets. Biochip systems have great promise to offer several advantages in size, performance, fabrication, analysis and production cost due to their integrated optical sensing microchip. The small sizes of the probes (microliter to nanoliter) minimize sample requirement and reduce reagent and waste requirement. Highly integrated systems lead to a reduction in noise and an increase in signal due to the improved efficiency of sample collection and the reduction of interfaces. The capability of large-scale production using low-cost integrated circuit (IC) technology is an important advantage. The assembly process of various components is made simple by integration of several elements on a single chip. For medical applications, this cost advantage will allow the development of extremely low cost, disposable biochips that can be used for in-home medical diagnostics of diseases without the need of sending samples to a laboratory for analysis.
Bioreceptors are the key to specificity for biosensor technologies. They are responsible for binding the analyte of interest to the sensor for the measurement. These bioreceptors can take many forms and the different bioreceptors that have been used are as numerous as the different analytes that have been monitored using biosensors. Current receptor technology based on enzyme-substrate interactions, nucleic acid methods, antibody-antigen interactions are highly specific. Broad spectrum detection of analytes is not possible using these
96 |
MIHRIMAH OZKAN ET AL. |
5 m
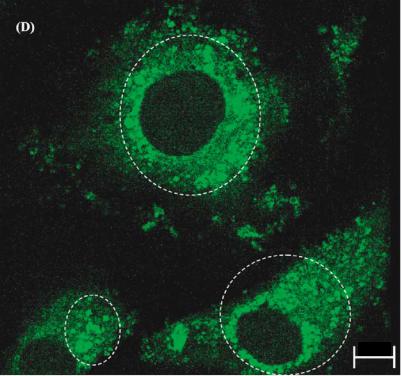
FIGURE 3.16(D). Physiological effect of EDTA at 280 ppm on osteoblasts. The cells are stained with fluo-3 for visualizing free calcium ion transients. The circled areas in the confocal optical micrograph show the presence of calcium bundles in the cytoplasm after exposure to fluo-3.
techniques. Moreover these methods are laboratory bound making them difficult to compartmentalize and commercialize other than for very specific applications as mentioned in previous sections. Thus there is a need for a different genre of sensors that have the broad based identification capability along with the associated advantages of biochip technology. Cell based biosensors offer this dual capability.
Cell-based biosensors constitute a promising field that has numerous applications ranging from pharmaceutical screening to environmental monitoring. Cells provide an array of naturally evolved receptors and pathways that can respond to an analyte in a physiologically relevant manner. Enzymes, receptors, channels, and other signaling proteins that may be targets of an analyte are maintained and, as necessary, regenerated by the molecular machinery present in cells. The array of signaling systems characteristic of cell-based sensors yields generic sensitivity that is a distinguishing feature in comparison to other molecular biosensor approaches. In addition, cell-based sensors offer an advantage of constituting a function-based assay that can yield insight into the physiologic action of an analyte of interest. Three important issues that constitute barriers for the use of cell-based sensors have been presented and discussed. There are certainly other areas that will require attention, as cell-based sensors move from the laboratory environment; namely, cell delivery and/or
MICROARRAY AND FLUIDIC CHIP FOR EXTRACELLULAR SENSING |
97 |
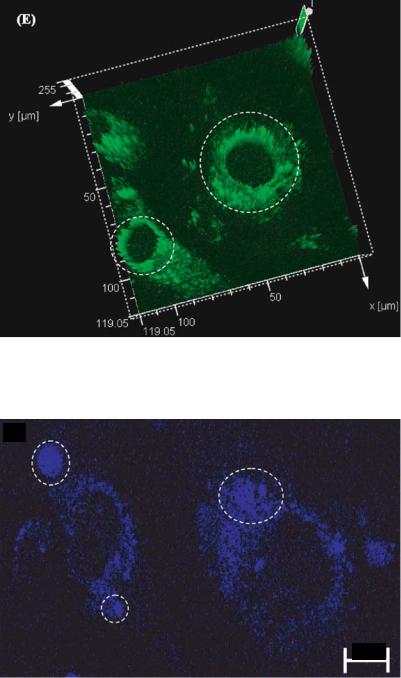
FIGURE 3.16(E). Three dimensional vector representation of the fluorescence intensity. The circled areas represent the high intensity peaks. The high intensity peaks are distributed uniformly throughout the cytoplasm. These areas correspond to the areas of localization of the free calcium ions released into the osteoblast’s cytoplasm due to the exposure to EDTA.
(F)
1 m
FIGURE 3.16(F). Single osteoblasts stained with Hoechst 33382 stain after being exposed to EDTA at 280 ppm concentration for a period of one sensing cycle. The circled areas indicate the regions of fragmentation of the nucleic acid. This behavior correlates with the results obtained from large cell populations after exposure to EDTA.
98 |
MIHRIMAH OZKAN ET AL. |
preservation technologies. As further progress is made to address fundamental challenges, cell-based biosensors and related cellular function based assays will undoubtedly become increasingly important and useful. It is of popular belief that such function-based assays will become an indispensable tool for monitoring in environmental, medical, and defense applications. Strategies relying on a single population of excitable cells appear most well suited for measurements of acute and direct effects of receptor agonist/antagonists. Compounds that fall within this category include ion channel modulators, metals, ligand–receptor blockers, and neurotransmitters. In fact, the detection of acute and direct effects of compounds may be sufficient and relevant for certain operational situations, such as a battlefield environment or the floor of an assembly plant, where cognitive function is absolutely critical. The prospect of detecting all physiologically active analytes using a single cell or tissue type is improbable. It is possible that particular analytes may undergo biotransformation, resulting in a secondary or tertiary compound of substantial physiologic effect. In spite of that drawback single cell based sensors are highly reliable. This has been shown by the newly developed single cell based sensing technique. This technique functions on the principle of integrating a fundamental biological tool like dielectrophoresis to biochip technology. Single cell arrays of the same biological state and differentiation can be developed using this method. Simultaneous sensing can be achieved, which reduces false alarms. Unique identification tags have been generated for identifying specific chemical analytes using this technique. These are known as Signature Patterns. The greatest advantage of this technique is its high sensitivity and speed of response. Chemical analytes of concentrations in the order of parts per billion have been detected. To determine the veracity and reliability of the sensor simultaneous fluorescence detection techniques have also been implemented at the detection limit obtained from the single cell based sensor. The physiological behavior corroborates the sensing. This establishes the viability of this technique for potential commercial implementation.
Finally, the threat of biological weapons has become a major concern to both the civilian and military populations [51]. All of the present biological warfare and environmental agent rapid detection systems, in field use or under prototype development, rely on structural recognition approaches to identify anticipated agents. Cell based sensor technology utilizing biochip capability can be thought to be one potentially reliable solution.
REFERENCES
[1]D. Aga. Environmental immunoassays: alternative techniques for soil and water analysis. In D. Aga, and E.M. Thurman (eds.), Immunochemical Technology for Environmental Applications, vol. 657, American Chemical Society, p. 212, 1997.
[2]B.M. Applegate, S.R. Kermeyer, and G.S. Sayler. Appl. Environ. Microbiol., 64:2730, 1998.
[3]J.Z. Bao, C.C. Davis, and R.E. Schmukler. Biophys. J., 61:1427, 1992.
[4]M.F. Bear, C.W. Barry, and M.A. Paradiso. Neuroscience: Exploring the Brain, 2nd ed., Lippincott, Williams and Wilkins, Baltimore, MD, p.147, 1999.
[5]F.F Becker, X.B. Wang, Y. Huang, R. Pethig, J. Vykoukal, and P.R.C. Gascoyne. J. Phys. D. Appl. Phys. 27:2659, 1994.
[6]S. Belkin, D.R. Smulski, S. Dadon, A.C. Vollmer, T.K. Van Dyk, and R.A. Larossa. Water Res., 31:3009, 1997.
[7]L. Bousse. Sens. Actu. B, 34:270, 1996.
[8]J.J. Broz, S.J. Simske, and A.R. Greenberg. J. Biomech., 28(11):1357, 1995.
MICROARRAY AND FLUIDIC CHIP FOR EXTRACELLULAR SENSING |
99 |
[9]L.I. Bruijn, M.K. Houseweart, S. Kato, K.L. Anderson, S.D. Anderson, E. Ohama, A.G. Reaume, R.W. Scott, and D.W. Cleveland. Science, 281:1851, 1998.
[10]R.S. Burlage, A.V. Palumbo, A. Heitzer, and G. Sayler. Appl. Microbiol. Biotechnol., 45:731, 1994.
[11]J.P.H. Burt, R. Pethig, M.S. Talary, and J.A. Tame. International Progress in Engineered Precision, Elseveir, Netherlands, p. 476, 1995.
[12]M.P. Byfield and R.A. Abuknesha. Biosens. Bioelectron., 9:373, 1994.
[13]L. Campanella, G. Favero, D. Mastrofini, and M. Tomasetti. J. Pharm. Biomed. Anal., 14:1007, 1996.
[14]D.W. Chan and T.D. Yager. Chromosomal, 107(1):39, 1998.
[15]J.C. Chang, G.J. Brewer, and B.C. Wheeler. J. Biomed. Microdev., 2(4):245, 2000.
[16]K. Carlson, B.S. Jortner, and M. Ehrich. Toxicol. Appl. Pharmacol., 168(2):102, 2000.
[17]D. Charych, Q. Cheng, A. Reichert, G. Kuziemko, M. Stroh, J.O. Nagy, W. Spevak, and R.C. Stevens. Chem. Biol., 3:113, 1996.
[18]H. Chen, D. Hayakawa, S. Emura, Y. Ozawa, H. Taguchi, R. Yano, and S. Shoumura. Histol. Histopathol., 6(3):763, 2001.
[19]P. Connolly, G.R. Moores, W. Monaghan, J. Shen, S. Britland, and P. Clark. Sens. Actu., B6:113, 1992.
[20]D. Cooke and R. O’Kennedy. Anal. Biochem., 274:188, 1999.
[21]B.A. Cornell, V.L. Braach Maksvytis, L.G. King, P.D. Osman, B. Raguse, L. Wieczorek, and R.J. Pace. Nature, 387:580, 1997.
[22]J. Csicsvari, D.A. Henze, B. Jamieson, K.D. Harris, A. Sirota, P. Bartho, K.D. Wise, and G. Buzsaki. J. Neurophysiol., 90:1314, 2003.
[23]K.W. Dunn, S. Mayor, J.N. Myers, and F.R. Maxfield. FASEB J., 8:573, 1994.
[24]C.F. Edman, D.E. Raymond, D.J. Wu, E.G. Tu, R.G.Sosnowski, W.F. Butler, M. Nerenberg, and M.J. Heller.
Nucleic Acids Res., 25:4907, 1997.
[25]R. Ehret, W. Baumann, M. Brischwein, A. Schwinde, K. Stegbauer, and B. Wolf. Biosens. Bioelectron., 12:29, 1997.
[26]S. Ekelund, P. Nygren, and R. Larsson. Anti-Cancer Drugs, 9:531, 1998.
[27]J.W. Ellwart and P. Dormer. Cytometry, 11:239, 1990.
[28]M. Emgard, K. Blomgren, and P. Brundin. Neuroscience, 115(4):1177, 2002.
[29]G.A. Evtugyn, E.P. Rizaeva, E.E. Stoikova, V.Z. Latipova, and H.C. Budnikov. Electroanalysis, 9:1, 1997.
[30]P.B. Fernandes. Curr. Opin. Chem. Biol., 2:597, 1998.
[31]F. Foch-Anderson and P. D’Orazio. Clin. Chem., 44:655, 1998.
[32]G. Fuhr, R. Hagedorn, T. Muller,¨ W. Benecke, B. Wagner, and J. Gisma. Stud. Biophys., 120:79, 1991.
[33]G. Fuhr, H. Glasser, T. Muller, and T. Schnelle. Biochim. Biophys. Acta, 1201:353, 1994.
[34]G. Fuhr, T.Muller,¨ T. Schnelle, R. Hagedorn, A. Voigt, S. Fiedler, W.M. Arnold, U. Zimmermann, B. Wagner, and A. Heuberger. Naturwissenschaften, 81:528, 1994.
[35]G. Fuhr and S.G. Shirley. J. Micromech. Microeng., 5:77, 1995.
[36]G. Fuhr, A. Voigt, T. Muller,¨ B. Wagner, K. Reimer, and T. Lisec. Sens. Actu. B, 26–27:468, 1995.
[37]I. Giaever and C.R. Keese. IEEE Trans. Biomed. Eng., 33:242, 1986.
[38]I. Giaever and C.R. Keese. Proc. Natl. Acad. Sci. USA, 88:7896, 1991.
[39]W. Gopel, A comprehensive survey, of sensors, Trends in sensor technology/sensor markets, Elseiver, Netherlands, p. 295, 1995.
[40]K.A. Giuliano, R.L. DeBiasio, R.T. Dunlay, A. Gough, J.M. Volosky, J. Zock, G.N. Pavlakis, and D.L. Taylor. J. Biomol. Screening, 2:249, 1997.
[41]C.R. Graham, D. Leslie, and D.J. Squirrell. Biosens. Bioelectron., 7:487, 1992.
[42]G.W. Gross, W. Wen, and J. Lin. J. Neurosci. Meth., 15:243, 1985.
[43]G.W. Gross, B.K. Rhoades, and R. Jordan. Sens. Actu. B, 6:1, 1992.
[44]G.W. Gross, B.K. Rhoades, D.L. Reust, F.U. Schwalm. J. Neurosci. Meth., 50:131, 1993.
[45]G.W. Gross. Internal dynamics of randomized mammalian neuronal networks in culture. In D.A. Stenger and T.M. McKenna (eds.), Enabling Technologies for Cultured Neuronal Networks, San Diego: Academic, San Diego, p. 277, 1994.
[46]G.W. Gross, B.K. Rhoades, H.M.E. Azzazy, and M.C. Wu. Biosens. Bioelectron., 10:553, 1995.
[47]L. Griscom, P. Degenaar, B. LePioufle, E. Tamiya, and H. Fujita. Sens. Actu. B, 83(1-3):15, 2002.
[48]E.A.H. Hall. Biosensors, Prentice Hall, Englewood Cliffs, New Jersey, 1991.
[49]J.M. Hall, J. More Smith, J.V. Bannister, and I.J. Higgins. Biochem. Mol. Biol. Int., 32:21, 1994.
[50]C.M. Hanbury, W.G. Miller, and R.B. Harris. Clin. Chem., 3:2128, 1997.
100 |
MIHRIMAH OZKAN ET AL. |
[51]D.A. Henderson. Science, 283:1279, 1999.
[52]A.W. Hendricson, M.P. Thomas, M.J. Lippmann, and R.A. Morrisett. J. Pharmacol. Exp. Ther., 307(2):550, 2003.
[53]T. Henning, M. Brischwein, W. Baumann, R. Ehret, I. Freund, R. Kammerer, M. Lehmann, A. Schwinde, and B. Wolf. Anticancer Drugs., 12(1):21, 2001.
[54]T.J. Heppner and J.F. Fiekers. Brain Res., 563:303, 1991.
[55]T.J. Heppner and J.F. Fiekers. Comp. Biochem. Physiol., 102C:335, 1992.
[56]H.R. Hoogenboom, A.P. de Bruine, S.E. Hufton, R.M. Hoet, J.W. Arends, and R.C. Roovers. Immunotechnology, 4:1, 1998.
[57]X. Hu, I.M. Johansson, T. Brannstrom, T. Olsson, and P. Wester. Acta Neuropathol., 104(5):462, 2002.
[58]A. Hulanicki, S. Glab, and F. Ingman. Pure Appl. Chem., 63:1247, 1991.
[59]C.K. Javawickreme, S.P. Javawickreme, and M.R. Lerner. Methods Mol. Biol., 87:119, 1998.
[60]M. Jenker, B. Muller, and P. Fromherz. Biol. Cybernetics., 84:239, 2001.
[61]T.B. Jones. Electromechanical Properties of Particles, Cambridge University Press, UK, p. 5, 1995.
[62]T.B. Jones and M. Washizu. J. Electrostat., 37:121, 1996.
[63]H. Kamioka, E. Maeda, Y. Jimbo, H.P.C. Robinson, and A. Kawana. Neurosci. Lett., 206:109, 1996.
[64]I.S. Kampa and P. Keffer. Clin. Chem., 44:884, 1998.
[65]I. Karube, K. Yokoyama, K. Sode, and E. Tamiya. Anal. Lett. 22(4):791, 1989.
[66]I. Karube, T. Matsunaga, S. Mitsuda, and S. Suzuki. Biotechnol. Bioeng., 19(10):1535, 1977.
[67]C.R. Keese and I. Giaever. IEEE Proc. Biomed. Eng., 12:500, 1990.
[68]J.M.H. King, P.M. DiGrazia, B. Applegate, R. Burlage, J. Sanseverino, P. Dunbar, F. Larimer, and G.S. Sayler. Science, 249:778, 1990.
[69]Y.I. Korpan, M.V. Gonchar, N.F. Starodub, A.A. Shul’ga, A.A. Sibirny, and A.V. El’skaya. Anal. Biochem., 215:216, 1993.
[70]V.G. Koveshnikov and V.S. Pikaliuk. Morfologiia, 104(3-4):364, 1993.
[71]C.M. Kurbacher, I.A Cree, and H.W. Bruckne. Anti-Cancer Drugs, 9:51, 1998.
[72]M.R. Lerner. Trends Neurosci., 17:142, 1994.
[73]Y.R. Li and J. Chu. Appl. Biochem. Biotechnol., 28-29:855, 1991.
[74]F.S. Ligler, T.L. Fare, K.D. Seib, J.W. Smuda, A. Singh, P. Ahl, M.E. Ayers, A. Dalziel, and P. Yager. Med. Instrum., 22:247, 1988.
[75]J. Liu, L. Bjo¨rnsson and B. Mattiasson. Biosens. Bioelectron., 14:883, 2000.
[76]M.P. Maher, J. Pine, J. Wright, and Y.C. Tai. J. Neurosci. Meth., 87:45, 1999.
[77]R.E. Maldve, T.A. Zhang, K. Ferrani-Kile, S.S. Schreiber, M.J. Lippmann, G.L. Snyder, A.A. Fienberg, S.W. Leslie, R.A. Gonzales, and R.A. Morrisett. Nat. Neurosci., 5:641, 2002.
[78]M. Malmquist. M. Biochem. Soc. T., 27:335, 1999.
[79]G.H. Markx, H. Talary, and R. Pethig. J. Biotechnology., 32:29, 1994.
[80]G.H. Markx and R. Pethig. Biotech. Bioeng., 45:337, 1995.
[81]S. Masuda, M. Washizu, and T. Nanba. IEEE Trans. Ind. Appl., 25:732, 1989.
[82]H.M. McConnell, J.C. Owicki, J.W. Parce, D.L. Miller, G.T. Baxter, H.G. Wada, and S. Pitchford. Science, 257:1906, 1992.
[83]M. Mehrvar, C. Bis, J.M. Scharer, M. Moo-Young, and J.H. Luong. Anal. Sci., 16:678, 2000.
[84]M. Meusel and T. Vering. Biosens. Bioelectron., 13:IX, 1998.
[85]K.M. Millan and S.R. Mikkelsen. Anal. Chem., 65:2317, 1993.
[86]C. Miller, Ion Channel Reconstitution, Plenum Press, New York, 1986.
[87]P. Mitra, C.R. Keese, and I. Giaever. Biotechniques, 1(4):504, 1991.
[88]T. Momiyama, Folia Pharmacol. Japon., 121(3):174, 2003.
[89]T. Muller,¨ A. Gerardino, T. Schnelle, S.G. Shirley, F. Bordoni, G. De Gasperis, R. Leoni, and G. Fuhr. J. Phys. D: Appl. Phys., 29:340, 1996.
[90]T. Muller,¨ S. Fiedler, T. Schnelle, K. Ludwig, H. Jung, and G. Fuhr. Biotechnol. Tech., 10:221, 1996.
[91]S.H. Nam, S.Y. Jung, C.M. Yoo, E.H. Ahn, and C.K. Suh. Yonsei. Med. J., 43(2):229, 2002.
[92]E. Neuwmann and A.E. Sowers. Electroporation and Electrofusion in Cell Biology, C.A. Jordan (ed.), Plenum Press, New York, NY, 1989.
[93]E. Noiri, Y. Hu, W.F. Bahou, C.R. Keese, I. Giaever, and M.S. Goligorsky. J. Biol. Chem., 272:1747, 1997.
[94]E.R. O’Connor, H.K. Kimelberg, C.R. Keese, and I. Giaever. Am. J. Physiol. 264(2 Pt 1):C471, 1993.
[95]A. Offenhausser, C. Sprossler, M. Matsuzawa, and W. Knoll. Biosens. Bioelectron., 12(8):819, 1997.
MICROARRAY AND FLUIDIC CHIP FOR EXTRACELLULAR SENSING |
101 |
[96]O. Orwar, K. Jardemark, I. Jacobson, A. Moscho, H.A. Fishman, R.H. Scheller, and R.N. Zare. Science, 272:1779, 1996.
[97]A. Ota and S. Ueda. Hybridoma, 17:471, 1998.
[98]B.M. Paddle. Biosens. Bioelectron., 11:1079, 1996.
[99]J.J. Pancrazio, S.A Gray, Y.S. Shubin, N. Kulagina, D.S. Cuttino, K.M. Shaffer, K. Eisemann, A. Curran,
B.Zim, G.W. Gross, and T.J. O’Shaughnessy. Biosens. Bioelectron., 18(11):1339, 2003.
[100]A. Parbhu, H. Lin, J. Thimm, and R. Lal. Peptides, 23(7):1265, 2002.
[101]R. Pethig. Dielectric and Electronic Properties of Biological Materials, J. Wiley and sons, United Kingdom, p. 186, 1979.
[102]R. Pethig and D.B. Kell. Phys. Med. Biol., 32:933, 1987.
[103]R. Pethig. Application of AC electrical fields to the manipulation and characterization of cells. In I. Karube (ed.), Automation in Biotechnology—Proc. 4t h Conf., Elsevier, Netherlands, p. 159, 1990.
[104]R. Pethig. Automation in Biotechnology, I. Kaurbe (ed.), Elsevier, Netherlands, p. 159, 1991.
[105]R.T. Piervincenzi, W.M. Reichert, and H.W. Hellinga. Biosens. Bioelectron., 13:305, 1988.
[106]P.A. Piunno, U.J. Krull, R.H. Hudson, M.J. Damha, and H. Cohen. Clin. Chem., 67:2635, 1995.
[107]H.A. Pohl, Dielectrophoresis, Cambridge University Press, United Kingdom, 1978.
[108]D. Pollard-Knight, E. Hawkins, D. Yeung, D.P. Pashby, M. Simpson, A. McDougall, P. Buckle, and S.A. Charles. Ann. Biol. Clin., 8:642, 1990.
[109]R.A. Potyrailo, R.C. Conrad, A.D. Ellington, and G.M. Hieftje. Anal. Chem., 70:3419–3425, 1998.
[110]S. Prasad, M. Yang, X. Zhang, C.S. Ozkan, and M. Ozkan. J. Biomed. Microdevices., 5(2):125, 2003.
[111]S. Prasad, X. Zhang, M. Yang, Y. Ni, V. Parpura, C.S. Ozkan, and M. Ozkan. J. Neurosci. Meth. (accepted)
[112]J.A.R. Price, J.P.H Burt, and R. Pethig. Biochim. Biophys. Acta, 964:221, 1988.
[113]B. Pucci, L. Bellincampi, M. Tafani, V. Masciullo, G. Melino, and A. Giordano. Exp. Cell Res., 252(1):134, 1999.
[114]K.R. Rogers and M. Mascini. Field. Anal. Chem. Technol., 2:317, 1998.
[115]K.R. Rogers. Biosens. Bioelectron., 10:533, 1995.
[116]C. Rosenmund and G.L. Westbrook. J. Physiol., 470:705, 1993.
[117]T.G. Ruardij, M.H. Goedbloed, and W.L.C. Rutten. Med. Biol. Eng. Comput. 41(2):227, 2003.
[118]J. Ruhe, R. Yano, J.S. Lee, P. Koberle, W. Knoll, and A. Offenhausser. J. Biomater. Sci. Polym. Ed., 10(8):859, 1999.
[119]F. Scheller and F. Schubert, Biosensors, Elsevier, Netherlands, 1992.
[120]M. Scholl, C. Sprossler, M. Denyer, M. Krause, K. Nakajima, A. Maelicke, W. Knoll, and A. Offenhausser.
J.Neurosci. Meth., 104:65, 2000.
[121]O. Selifonova, R.S. Burlage, and T. Barkay. Appl. Environ. Microbiol. 59:3083, 1993.
[122]G.J. Singh and B. Singh. Neurobehav. Toxicol. Teratol., 6(3):201, 1984.
[123]J. Singh, P. Khosala, and R.K. Srivastava. Ind. J. Pharmacology., 32:206, 2000.
[124]M.A. Smith, P.S. Herson, K. Lee, R.D. Pinnock, and M.L. Ashford. J. Physiol., 547(Pt 2):417, 2003.
[125]A. Steinstrasser, A. Schwarz, C. Alexander, R. Berberich, and M. Zimmer. Nuklearmedizin, 31(5):164, 1992.
[126]D.A. Stenger, D.H. Cribbs, and T.L. Fare. Biosens. Bioelectron., 6:425, 1991.
[127]M. Stephens, M.S. Talary, R. Pethig, A.K. Burnett, and K.I. Mills. Bone marrow Transplant., 18:777, 1996.
[128]A.K. Stout, H.M. Raphael, B.I. Kanterewicz, E. Klann, and I.J. Reynolds. Nature. Neurosci., 1(5):366, 1998.
[129]E. Sullivan, E.M. Tucker, and I.L. Dale. Methods Mol. Biol., 114:125, 1999.
[130]J.R. Subramaniam, W.E. Lyons, J. Liu, T.B. Bartnikas, J. Rothstein, D.L. Price, D.W. Cleveland, J.D. Gitlin, and P.C. Wong. Nature Neurosci., 5:301, 2002.
[131]R.M. Sutherland, C. D¨ahne, J.F. Place, and A.R. Ringrose. Clin. Chem., 30:1533, 1984a.
[132]R.M. Sutherland, C. D¨ahne, J.F. Place, and A.R. Ringrose. J. Immunol. Methods., 74:253, 1984b.
[133]T.C. Tan and Z.R. Qian. Sens. Acuta B, 40:65, 1997.
[134]T. Tanaka, Y. Taniguchi, K. Gotoh, R. Satoh, M. Inazu, and H. Ozawa. Bone, 14(2):117, 1993.
[135]Y. Taniguchi, T. Tanaka, K. Gotoh, R. Satoh, and M. Inazu. Calcif. Tissue Int., 53(2):122, 1993.
[136]D.R. Thevenot, K. Toth, R.A. Durst, and G.S. Wilson. Pure Appl. Chem., 71:2333, 1999.
[137]P. Thiebaud, L. Lauer, W. Knoll, and A. Offenhausser. Biosens. Bioelectron. 17(1-2):87, 2002.
[138]R.Y. Tsien. Trends Neurosci., 11:419, 1988.
[139]T. Vo-Dihn, B. Cullum, and B. Fresenius. J. Anal. Chem., 366:540, 2000.
