
- •Accessory and pseudo receptors: betaglycan, endoglin, cripto, and BAMBI
- •Betaglycan
- •Cripto
- •BAMBI
- •Downstream signalling: Drosophila, Caenorhabitidis, and Smad
- •Smad proteins have multiple roles in signal transduction
- •Receptor-regulated Smads 1, 2, 3, 5 and 8: receptor recognition
- •Cytoplasmic retention of receptor-regulated Smad proteins
- •Common mediator Smad4
- •Hetero-oligomeric complex formation
- •Smad–Smad complexes
- •Nuclear import and export
- •SMAD transcriptional complexes
- •Activation of gene expression
- •Repression of gene expression
- •A self-enabling response: repression of myc is prerequisite for expression of cell cycle inhibitors
- •The Smad linker region: hotspot for kinases and an E3-ligase
- •Smurf-mediated Smad degradation
- •Inhibitory Smads
- •BAMBI, a signal inhibitory pseudo receptor
- •Smad phosphatases
- •References
Signal Transduction
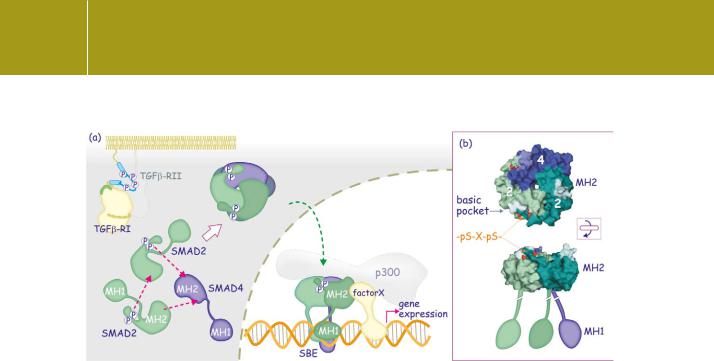
Fig 20.10 Smad activation and nuclear translocation.
(a) On phosphorylation of the C-terminal SxS motif, receptor-regulated Smads complex with each other and then with Smad4. The phosphoserines bind the basic pocket in the MH2 domain. The trimers enter the nucleus to bind DNA at the SBE. The complexes also bind other DNA-binding proteins and transcriptional cofactors (for instance p300). (b) Detail of a Smad2–Smad4 complex showing the interaction of the phosphoserines in the C-terminus with the basic pocket of the MH2 domain. The Smad proteins are viewed from two different angles to show the presumed orientation of the MH2 domain relative to the DNA-binding MH1 domain (1u7v51).
heterotrimers made up of two phospho-R-Smads and one Smad4 seem to be prevalent.51
Individual Smads also complex with Tcf/ -catenin or TIF1
Both Smads 2/3 and Smad4 also combine to form complexes with other DNA-binding proteins. Quite independently of TGF signalling, Smad4 forms a complex with Tcf and -catenin to induce expression of c-myc and the tight junction protein claudin-1 (Figure 20.11).53,54 Smads 2 and 3 bind TIF1 in competition with Smad4.55 This alternative complex comes into action during TGF -mediated differentiation of haematopoietic progenitors into red blood
cells, acting in parallel with the canonical TGF pathway, giving rise to a complex of Smads 2/3 with Smad4, which inhibits the proliferation of the progenitor cells.
Nuclear import and export
Migration of the Smads into the nucleus occurs both with and without
the intervention of nuclear importins. The hydrophobic corridor of the MH2 domain interacts directly with the FG repeat region on nucleoporins (see Figure 20.13), in this way obviating the nuclear transport receptors (importinsand - ).56 The Smads are then retained within the nucleus through the formation of hetero-oligomeric complexes that bind DNA and/or transcription
612

Signalling Through Receptor Serine/Threonine Kinases
Fig 20.11 A self-enabling response regulates expression of p21CIP1.
(a) Expression of the cyclin-dependent kinase inhibitor p21CIP1 is repressed by the transcription factors c-Myc and Miz, both expressed through the influence of -catenin/Tcf complexed with Smad4. (b) TGF inverses the situation. Smad3/Smad4 bind the TGF -inhibitory element in the c-myc promoter. With E2F4 (or -F5) and p107 they repress expression of c-myc. The c-Myc/Miz-mediated repression is lifted and a new transcriptional complex, comprising Smad3/Smad4, FoxO, and p300 drive expression of p21CIP1. After translation the inhibitor binds to CDK4–CyclinD and halts cell cycle progression. Regulation of the inhibitor p15INK4B is similar.
cofactors. Dephosphorylation of the SxS motif leads to dissociation and allows export back to the cytosol (see Figure 20.17).
SMAD transcriptional complexes
Smad hetero-trimers bind to DNA through their MH1 domains, but due to the weak interaction, they interact with other DNA-binding factors to achieve high affinity and selectivity.57 Furthermore, the sequence of the -hairpin that interacts with the base pairs that constitute the SBE is highly conserved (Figure 20.8). This suggests that it is not the DNA contact that provides selectivity for gene targeting between the different Smad proteins.58 Rather, the combinations of the Smads with their partners provide the necessary levels of specificity. Several cofactors have been identified, belonging to different families of DNA-binding proteins, that account for the great breadth of TGF transcriptional responses. Examples of complexes are presented in
613

Signal Transduction
Table 20.1. Important in all this is the recruitment of additional cofactors that either render the DNA accessible or inaccessible to RNA polymerase.
Activation of gene expression
In addition to all that has just been related, the heterotrimeric Smad complexes also interact with other DNA-binding proteins and transcriptional cofactors (see below) to form even bigger aggregates. These recruit histone acetyltransferases (HATs) such as p300 and CBP (see Figure 14.3, page 423)67,68 and, through interaction with the MH2 domains of Smads1–4, render the DNA accessible to RNA polymerases.
Repression of gene expression
About a quarter of all TGF responses in mammalian cells involve gene repression.69 Repressive complexes act in different ways. For instance, p107, a protein that resembles the retinoblastoma tumour suppressor, recruits histone methyltransferases to the promoter region to create silent heterochromatic regions.70 TGIF recruits, via the intermediate of CTBP1, the
histone deacetylation complex mSin3/HDAC and this leads to tightly wrapped DNA that is inaccessible to RNA polymerases.71
A self-enabling response: repression of myc is prerequisite for expression of cell cycle inhibitors
A common response to TGF in epithelial cells, inhibition of the cell division cycle, occurs by enhancing the expression of inhibitors of cyclin-dependent protein kinases. This is mediated through a self-enabling process in which Smad proteins first repress the expression of c-myc and then upregulate expression of the inhibitors. This has been studied in detail for p21Cip1 and p15INK4B.57,72 The c-myc promoter contains two Tcf-binding elements (TBE) that bind Tcf/LEF, -catenin, and Smad4 and drive gene expression. The c-myc promoter region also contains a degenerate Smad-binding sequence, TGF -inhibitory element (TIE) (Figure 20.11).73 In response to TGF , this binds Smads 3 and 4 combined with E2F4 (or E2F5). This complex then recruits the corepressor p107, which leads to effective silencing of c-myc.65 Since the promoter region of the two cell cycle inhibitors contains a c-Myc/Miz binding element that represses its transcription, loss of c-myc abrogates this negative control and leaves the way open for Smad-mediated activation of
transcription. This occurs through two promoter elements, SBE binding Smads 3 and 4 and FHBE which binds the forkhead transcription factor FoxO.53 Together these recruit p300 which opens the way for the RNA polymerase. Elevated levels of p21CIP1 and p15INK4b then inhibit the cell cycle through binding and inhibiting the kinase complex cyclinD–CDK4 that facilitates progression through the G1 phase.
614

Signalling Through Receptor Serine/Threonine Kinases
Table 20.1 Transcription factors that interact with Smad heterotrimeric complexes
Protein family |
Members |
Interaction |
Gene and function |
|
|
|
|
Forkhead family |
FoxH1 (FAST1) |
Smad2 |
induction of expression of Mix, leading to |
(winged helix |
|
Smad4 |
mesoderm induction in Xenopus in response |
proteins) |
|
|
to activin or nodal-like signals59 |
|
FoxO1, FoxO2, FoxO3 |
Smad3 |
induction of expression of p21CIP1 in epithelial |
|
|
Smad4 |
cells, leading to inhibition of the cell division |
|
|
|
cycle57 |
RUNX |
Runx1, -2, -3 (also |
Smad2 or -3 |
induction of expression of the constant |
|
known as Cbfa) |
Smad4 |
region of Ig in B cells60 |
|
|
|
repression of expression of osteocalcin, |
|
|
|
leading to the inhibition osteoblast |
|
|
|
differentiation61 |
Mix |
Mixer, Milk |
Smad3 |
restriction of Goosecoid expression to the |
Homeodomain |
|
Smad4 |
dorsal marginal zone of the early Xenopus |
|
|
|
gastrula embryo62 |
Zinc finger |
OAZ (ZNF423) (Olf-1 |
Smad1 |
induction of expression of vent2, a gene that |
proteins |
associated zinc |
Smad4 |
orchestrates ventral mesoderm induction in |
|
finger) |
|
Xenopus in response to BMP63 |
AP1 |
c-Jun, c-fos |
Smad3 |
induction of expression of junB, c-jun, |
|
|
Smad4 |
collagenase-1 (mmp1), interleukin-11 and |
|
|
|
plasminogen activator inhibitor-164 |
E2F/DP |
E2F4, E2F5 |
Smad3 |
repression of expression of c-myc by TGF in |
|
|
Smad4 |
epithelial cells, leading to cell cycle inhibition |
|
|
|
(through release of c-Myc-mediated |
|
|
|
repression of cell cycle inhibitors)65 |
ATF/CREB |
ATF3 |
Smad3 |
repression of expression of Id1, leading to cell |
|
|
Smad4 |
cycle inhibition of epithelial cells66 |
Smad proteins as integrators of signal pathways
Through their cooperativity and the multitude of their interactions, the Smads provide a versatile platform that provides many forms of cross-talk between signalling pathways. These are summarized in Figure 20.12 and in the following paragraphs.
•
•
The activation of STAT3 (page 525) by leukaemia inhibitor factor (LIF) enforces BMP-induced expression of GFAP. This way, STAT3 and Smad1 work together to drive differentiation of neural stem cells into astrocytes. p38 and JNK (page 350) contribute to TGF signalling through activation of ATF3 and c-Jun, both of which interact with Smad proteins. ATF3 and
615
