
- •Accessory and pseudo receptors: betaglycan, endoglin, cripto, and BAMBI
- •Betaglycan
- •Cripto
- •BAMBI
- •Downstream signalling: Drosophila, Caenorhabitidis, and Smad
- •Smad proteins have multiple roles in signal transduction
- •Receptor-regulated Smads 1, 2, 3, 5 and 8: receptor recognition
- •Cytoplasmic retention of receptor-regulated Smad proteins
- •Common mediator Smad4
- •Hetero-oligomeric complex formation
- •Smad–Smad complexes
- •Nuclear import and export
- •SMAD transcriptional complexes
- •Activation of gene expression
- •Repression of gene expression
- •A self-enabling response: repression of myc is prerequisite for expression of cell cycle inhibitors
- •The Smad linker region: hotspot for kinases and an E3-ligase
- •Smurf-mediated Smad degradation
- •Inhibitory Smads
- •BAMBI, a signal inhibitory pseudo receptor
- •Smad phosphatases
- •References

Signal Transduction
The nematode C. elegans responds to conditions of overcrowding and starvation by developmental arrest as a dauer (resilient, durable) larva. Screening of mutants having this phenotype revealed a number of genes (daf-1–4, etc.) of which daf-1 and daf-4 code for serine/threonine receptor protein kinases.34,35 daf-4 mutants are dauer-constitutive and the larvae are smaller than the wild types. Screening for mutants with similar phenotypes has revealed three more genes, sma-2–4,36 which act downstream of daf-4. Mad (fly) and Sma (worm) proteins are homologous and with the sequences to hand, eight human homologues coding for Smad proteins have been identified.14,37
Smad proteins have multiple roles in signal transduction
The Smad proteins have two regions of homology, MH1 and MH2, connected by a more divergent linker segment (Figure 20.8a). The N-terminal MH1, highly conserved in all Smads (except Smads 6 and 7), binds to DNA at the Smad binding element (though, curiously, the most abundant splice form of Smad2 carries an insert that blocks binding to DNA). MH1 also interacts with transcription factors such as Jun, ATF3, Sp1, and Runx. The linker constitutes a flexible segment that is a hotspot of phosphorylation sites and operates in the integration of several signalling pathways. It also contains a PPxY motif that acts as an E3-ubiquitin ligase binding site. In Smad4 the linker contains a nuclear export signal (NES) (Figure 20.8b).
The MH2 domain, a particularly versatile protein-interacting module, is conserved in all Smad proteins. A set of adjoining hydrophobic patches (the hydrophobic corridor) mediates interactions with cytoplasmic retention proteins such as SARA, nuclear pore proteins (NUP214) and with a number of transcription cofactors. The MH2 domains of Smads 1, 2, 3, 5 and 8 carry a
conserved C-terminal SxS motif, the substrate of T R-I. When phosphorylated, this interacts with a basic pocket in MH2 to provoke the assembly of heterooligomeric complexes (see below).
Smad proteins have different functions, controlled through their selective interactions with T R-I and with each other38 (Figures 20.9 and 20.10). On the basis of their sequences and functions they are divided into three groups: (i) receptor-regulated Smads 1, 2, 3, 5, and 8; (ii) common mediator Smads 4 and 3; inhibitory Smads (6 and 7). Below we elaborate further on their structure and function.
Receptor-regulated Smads 1, 2, 3, 5 and 8: receptor recognition
The receptor-regulated Smads are phosphorylated by the activated T RI receptors. Short structural elements in T RI and in the Smads determine the specificity of interaction: the exposed L45 loop in the type I receptor kinase
608

Fig 20.8 Molecular structure and domain architecture of Smad proteins.
(a) Receptor-regulated Smad proteins have three distinct segments, a conserved MH1 domain, a more divergent linker region, and a highly conserved MH2 domain. MH1 is stabilized by Zn2 (yellow sphere) and contains a -hairpin that binds the Smad-binding element (SBE). The MH2 domain contains a C-terminal SxS motif that is phosphorylated by the type I receptor. It also has an L3 loop and basic pocket which interact with L45 and the phosphorylated GS domain of the receptor. It exposes a hydrophobic corridor which is the site of attachment of numerous proteins among which is SARA. It also interacts with
a large number of other components including T R-1, nuclear pore proteins, coactivators and repressors, and DNA binding cofactors. It is involved in Smad oligomerization (1ozj,39 1 khx40). (b) Smad transcription factors are subdivided in three groups based on functional and structural criteria. Receptor-regulated Smad proteins contain the SxS motif which is phosphorylated by type I receptors. They contain a NUP-binding motif as well as a PPxY motif with which they interact with the E3-ubiquitin ligase Smurf. The common mediator Smad4 forms complexes with all receptor-regulated Smad proteins interacting with their phosphorylated SxS motif. It lacks the L3 loop and does not interact with type 1 receptors. Smads 6 and 7 lack a functional MH1 domain as well as the SxS motif. They prevent TGF signalling. They too bind Smurf through a PPxY motif in the linker region. All Smad proteins contain numerous sites that are phosphorylated by serine/threonine protein kinases (just a few are indicated).
609
Signal Transduction
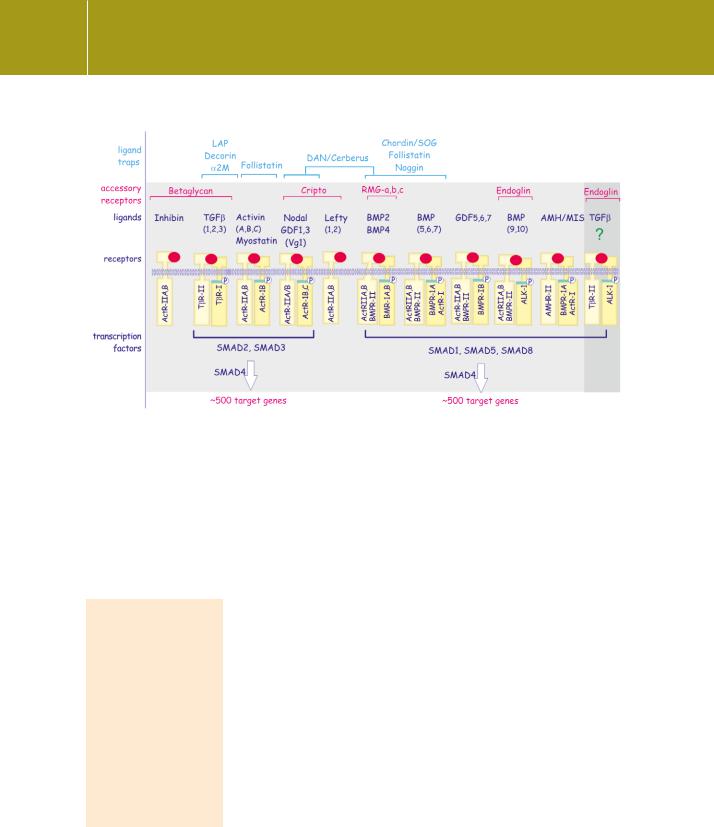
Fig 20.9 Ligands and their traps of the TGF family.
Traps are proteins that trap ligand or block its access to the receptor. Accessory receptors facilitate ligand binding and receptor complex formation. The different receptor combinations can be divided into two groups: the TGF /activin group recruits Smad-2 and 3, whereas the BMP group recruits Smad 1, 5, and 8. The type II receptors phosphorylate and activate the type I receptors. Unfortunately receptor nomenclature remains unsettled; ActR-1 = ALK-2, ActR-1B = ALK-4, ActR-1C = ALK-7. The combination in the shaded column on the right is not firmly established.
The MH2 domain qualifies as a phosphoserine-binding domain and offers yet another means by which receptor signalling complexes are formed. Structurally, it shares striking homology with the forkhead-associated domain (FHA) but not with 14-3-3, both of which also recognize phosphoserine/threonine residues.43
domain interacting with the L3 loop in the MH2 domain of the Smads (see Figures 20.3 and 20.8). By exchanging selected residues in these domains it is possible to switch the signalling specificity of the TGF and BMP pathways.41,42 Thus, the T R-I (ALK-5) and the nodal/activin type I receptors (ALK-4 and ALK- 7) recognize Smads 2 and 3, whereas ALK-1, -2, -3 and -6 recognize Smads 1,
5 and 8 (Figure 20.9). In this way, different ligands, binding different receptor combinations, provide different signals through the recruitment of specific combinations of Smad proteins.
Whereas the interaction between loops L45 and L3 determines specificity, phosphorylation of the GS domain acts as the on/off signal, roughly equivalent to phosphotyrosine binding to SH2or PTB domain containing proteins (see page 768). The phosphorylated GS domain is recognized
by a basic pocket in the Smad proteins, adjacent to the L3 loop (Figure 20.8b). Serine phosphorylation of the SxS motif in the MH2 domain causes a structural alteration that decreases affinity of the Smads for cytoplasmic anchors, increases their affinity for nuclear pore proteins and allows them to bind each other and to Smad4 (Figure 20.10).
610
