
- •Modular structure of proteins
- •Structural domains
- •The evolution and shuffling of domains
- •Sequence homology and the acquisition of function
- •Domain function
- •Catalytic domains
- •Protein interaction domains
- •The inventory of domains
- •Detection
- •Classification
- •Examples of domains with roles in signalling
- •Domains that bind oligopeptide motifs
- •SH2 domains
- •PTB domains
- •SH3 domains
- •Phosphoinositide-binding domains
- •PH domains
- •Other phosphoinositide-binding domains
- •Polypeptide modules that bind Ca2
- •Calcium-binding motifs and domains
- •The EF-hand motif
- •C2 domains
- •Zinc finger domains
- •Protein kinase domains
- •Protein kinases share a common domain
- •Structural elements that regulate kinase activity
- •References

Signal Transduction
such as this cannot make contact with more than four bases. Recognition of a segment of DNA long enough to represent a transcriptional site generally requires three or more such domains, separated by linkers.
In other families, Zn2 ions are coordinated by sets of four cysteine side chains (C2C2). This occurs in GATA-type zinc fingers (a group that takes its name from the erythroid transcription factor GATA-1, so-called because it binds to the DNA sequence GATA). Again, for this protein to bind with high affinity at a transcription site, more than one C2C2 module is required, in this case a tandem pair separated by a linker. The attachment of C2C2 fingers to DNA is illustrated in Figure 10.8, page 286 for the DNA-binding domain of the glucocorticoid receptor. The tandem C2C2 fingers are separated by a linking sequence, enabling the receptor to bind to the two half-sites on DNA.
A rather different three-dimensional structure is exhibited by the C2C2 finger that occurs in the lipid-binding FYVE domain. In the tandem arrangements of zinc fingers that we have considered so far, the N-terminal C2C2 binds one
Zn2 while the C-terminal C2C2 binds the other. In the FYVE domain, alternate pairs of cysteines bind to each Zn2 . This so-called cross-braced conformation is illustrated in Figure 24.5 (compare Figure 24.8).
Another example of the cross-brace occurs in the zinc finger domain that lies within the N-terminal region of conventional PKC. This is the regulatory C1 region that binds the activator, diacylglycerol. It contains two cysteine-rich zinc-binding domains, termed C1A and C1B (see Figure 9.9 page 256). Each of these can bind two Zn2 ions, each complexed by three cysteines and one histidine. Once again, alternate pairs of residues contribute to each site, so stabilizing the tertiary structure.46,47
Other zinc finger domains include the RING fingers involved in the ubiquitylation of proteins and in the consequent formation of multiprotein complexes (see figure 15.12 page 468).
Protein kinase domains
Protein kinases share a common domain
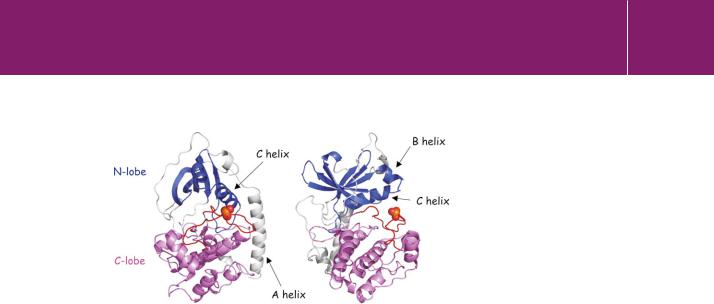
The catalytic activity of protein kinases (whether serine/threonine specific or tyrosine specific) is confined to a structurally conserved protein kinase domain. The basic architecture of the kinase domain, as typified by the catalytic subunit of PKA, is illustrated in Figure 24.9. The single polypeptide chain folds to form two closely apposed lobes, with an ATP binding site situated in the cleft formed between them. There is also an N-terminal - helical chain (the A helix) that makes contact with the surface of both lobes.
The N-terminal lobe, the smaller of the two, possesses 100 amino acids. Although it is myristoylated at its N-terminus, there is no evidence that this group is free to associate with membranes. Instead, it occupies a pocket and
782
Protein Domains and Signal Transduction
FIG 24.9 Protein kinase domain structure.
Two views of the catalytic subunit of porcine PKA. The structure on the right has been rotated 90° about a vertical axis. The small N-terminal lobe is coloured blue and the large C-terminal lobe magenta. The activation segment is coloured red and the phosphate group on T197 is depicted as spheres. An N-terminal myristoyl group is not shown (1cdk.pdb49).
provides structural stability, helping to keep the A-helix in contact with the larger lobe. The principal feature of the small lobe is an antiparallel -sheet. There are also two -helical chains, the B- and C-helices. The C-terminal lobe consists of 200 residues and it possesses mostly -helical structure, arranged around a stable four-helix bundle. Like other protein kinases, PKA has a central chain of residues called the activation segment or loop (Figure 24.9). The conformation of this segment and in many cases, the phosphorylation of a key residue (T197 in PKA) is critical for catalysis.48
Structural elements that regulate kinase activity
The principal components of the catalytic site are shown in Figure 24.10. In broad terms, the N-lobe binds ATP, while the C-lobe binds substrate and enables catalysis, though for ATP to bind in the correct orientation in the cleft, it makes contact with residues on both lobes. Two Mg2 ions are also bound (for clarity, these are not shown). The residues on the N-lobe that align the nucleotide include the main chain nitrogens of the glycine-rich loop between-strands 1 and 2, and the side chain of a lysine (K72) on -strand 3, that interacts in turn with a glutamate (E91) on the C-helix. The correct location of ATP is very sensitive to the positioning of the C-helix.
In the active conformation of the kinase, T197 is phosphorylated and makes contact with positively charged residues on both the small and large lobes. These interactions effectively seal the cleft and prepare for catalysis by neutralizing the charge on R165 in the catalytic loop, adjacent to D166, the key catalytic residue. With ATP in place, the recognition and binding of a consensus motif on the target protein can proceed. For PKA, the sequence of the motif is Rxx[T/S]h (where h indicates an amino acid with a hydrophobic
783

Signal Transduction
Fig 24.10 Catalytic core of PKA.
In this stereoscopic view (page xxv) of the catalytic site, the key chains are the glycine-rich loop (orange, top left of structure), the C-helix (magenta), the activation segment (red) and the catalytic loop (cyan and blue). Within these, important side chains include those of K72 in -strand 3 (green dotted spheres, top left), E91 in the C-helix
(magenta dotted spheres), and R165 in the catalytic loop (blue dotted spheres, bottom right). Phosphorylated T197 in the activation segment (yellow chain, phosphates not shown) interacts with residues in both lobes, including R165, neutralizing positive charges. D166 (cyan dotted spheres) is the catalytic aspartate. For experimental reasons, the bound nucleotide (spheres) is the ATP analogue, adenylyl imidodiphosphate (1cdk.pdb49).
Fig 24.11 Substrate binding at the catalytic site of PKA.
A substrate is mimicked by residues 5–24 of PKI (a PKA inhibitor; black). The key chains in the cleft and the nucleotide are depicted and coloured as in Figure 24.10. The view on the right has been rotated 90° about a vertical axis (1cdk.pdb49).
side chain). Binding of substrate in the correct orientation is essential. The serine or threonine hydroxyl group within the motif must be accurately aligned with the terminal phosphate of ATP, as illustrated in Figure 24.11. The achievement of this alignment depends critically on the conformation of the activation segment, and in this context the contacts made by pT197 with residues on both lobes are particularly important.
784

Protein Domains and Signal Transduction
Table 24.3 Activation segment phosphorylation
Phosphorylated in the activation segment |
Not phosphorylated in the activation |
||
|
|
segment |
|
|
|
|
|
Cyclic AMP-dependent kinase |
PKA |
Phosphorylase kinase |
PHK |
|
|
|
|
Protein kinase B |
PKB , , |
Casein kinase I |
CKI |
|
|
|
|
Protein kinase C |
PKC , II |
EGF receptor |
EGFR |
|
|
|
|
Cyclin-dependent kinase |
CDK1 CDK2 CDK7 |
C-terminal Src kinase |
Csk |
|
|
|
|
MAP kinase |
ERK1/2 |
Ca2 /calmodulin kinase |
CaMKII |
MAP kinase kinase |
MEK1 |
Myosin light chain kinase |
MLCK |
|
|
|
|
Raf1 kinase |
Raf1 |
|
|
|
|
|
|
Ca2 /calmodulin kinase |
CaMKI |
|
|
|
|
|
|
Insulin-stimulated kinase |
ISPK |
|
|
|
|
|
|
Glycogen synthase kinase |
GSK3 |
|
|
|
|
|
|
Insulin receptor kinase |
IRK |
|
|
|
|
|
|
PDGF receptor |
PDGFR |
|
|
|
|
|
|
c-Src family |
Src, Yes, Fyn, Fgr, |
|
|
|
Lyn, Lck, Blk |
|
|
|
|
|
|
Many other kinases are also phosphorylated in their activation segments at positions equivalent to T197 in PKA (see Table 24.3). These include isoforms of PKC, ERKs 1 and 2, MEK1, the cyclin-dependent kinases, CDK2 and CDK7, and the Src family protein tyrosine kinases. Although such
phosphorylations are conditional for the activation of these and other kinases, they are not all achieved through autophosphorylation. For example, in PKC, ERK, and the cyclin-dependent kinases, the sequence of the target motif in the activation segment does not match the consensus sequence recognized by the kinase. This implies a requirement for another upstream kinase
(for example phosphoinositide-dependent protein kinase 1 for PKC, see page 258).
Other protein kinases have no need for phosphorylation in their activation segments. For instance, on phosphorylase kinase, the side chain carboxyl groups of E182 fulfil a function similar to the phosphate groups of T197 of PKA, neutralizing the positive charge of the catalytic R148 adjacent to D149. (These residues correspond to R165 and D166 in PKA.) Nor does activation segment phosphorylation occur in those kinases in which the catalytic aspartate is preceded by a non-polar residue instead of arginine. Examples are twitchin
785

Signal Transduction
|
kinase and MLCK (myosin light chain kinase). In spite of possessing a tyrosine |
|
|
phosphorylation site in its activation segment, the EGF receptor kinase (ErbB1) |
|
|
is not activated by phosphorylation: instead this is achieved through allosteric |
|
|
interactions between the two catalytic domains (see Figure 12.4, page 321). |
|
List of abbreviations |
|
|
|
|
|
APAF |
apoptosis activating factor 1 |
|
|
|
|
Akt |
see PKB |
|
|
|
|
Arf |
GTPase (involved in vesicular transport) |
|
|
|
|
ARK |
-adrenergic receptor kinase (GRK2) |
|
|
|
|
Btk |
Bruton’s tyrosine kinase |
|
|
|
|
CaM |
calmodulin |
|
|
|
|
Caspase |
cysteinyl aspartate-specific protease |
|
|
|
|
CBP |
histone acetyltransferase, creb-binding protein |
|
|
|
|
DAG |
diacylglycerol |
|
|
|
|
Dbl |
Rho GEF (diffuse B cell lymphoma oncogene) |
|
|
|
|
EEA1 |
early endosome antigen 1 |
|
|
|
|
FADD |
Fas associated protein with death domain |
|
|
|
|
G , G |
and subunits of G proteins |
|
|
|
|
GEF |
guanine nucleotide exchange factor |
|
|
|
|
GCAP |
guanylate cyclase activating protein |
|
|
|
|
Grb2 |
SH2-SH3 adaptor ( growth factor receptor-bound protein 2) |
|
|
|
|
GRK |
G protein coupled receptor kinase (GRK2 ARK) |
|
|
|
|
GRP1 |
Arf GEF |
|
|
|
|
HP1 |
non-histone chromosomal protein (heterochromatin protein 1) |
|
|
|
|
InaD |
links PKC to PLC in Drosophila photoreceptors |
|
|
|
|
IRS1, IRS2 |
insulin receptor substrate 1 and 2 |
|
|
|
|
Nck |
SH2-SH3 adaptor |
|
|
|
|
p300 |
histone acetyltransferase (related to CBP) |
|
|
|
|
p40phox, p47phox |
NADPH oxidase subunits |
|
PI3K-p85 |
P85 subunit of PI 3-kinase |
|
|
|
|
|
Continued |
|
786
