
CST 2015 / MAPK_ERK_Growth
.pdf
CELL SIGNALING TECHNOLOGY |
www.cellsignal.com |
MAPK/Erk in Growth and Differentiation |
|
RTKs |
Integrins |
Ion Channels |
RTKs |
[Ca2+] |
|
[Ca2+]
MEK1
LAMTOR2
MP1
Erk1
Late Endosome
|
FRS2 |
GRB2 |
SOS |
Ras |
|
|
|
||
PLCγ |
Shc IRS |
|
Spry |
|
|
|
|
||
|
Src |
|
|
|
|
PYK2 |
|
Spred |
|
|
|
|
|
|
SOS |
GRB2 |
CAS |
Tal |
Rap1 |
|
|
RACK1 |
|||
PI3K |
|
Fyn |
Pax FAK |
C3G |
|
|
|
|
EPAC |
||
|
|
|
|
|
|
PAK |
Rac |
[cAMP] |
Raptor |
||
|
|||||
|
|
c-Raf |
|
PKA |
|
AMPK |
||||
PKC |
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
||
|
|
c-Raf B-Raf |
B-Raf |
|
|
|
|
|||
|
|
|
|
|
LKB1 |
|||||
|
|
|
|
Heterodimer |
14-3-3 |
|
|
|||
|
Tpl2/Cot1 |
|
|
|
PD98059 |
|
|
|
|
|
|
|
|
|
U0126 |
|
|
|
|
||
|
IMP |
|
|
|
AZD6244 |
|
|
|
|
|
Ion Channels, |
|
|
|
PP1/ |
FMK |
|||||
|
C-TAK1 |
|
KSR |
|
PP2At |
|||||
Receptors |
|
|
|
|
SL0101 |
|||||
cPLA2 |
|
|
1/2 |
|
SOS |
BI-D1870 |
||||
|
|
|
||||||||
|
|
|
MEK1/2 |
|
|
|
|
|||
|
|
|
|
Bim |
|
|
|
|||
|
|
|
|
|
||||||
Translation |
MNK1/2 |
|
|
Erk1/2 |
|
p90RSK |
||||
Control |
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
||||
CRK

mTOR Signaling
rpS6 TCPβ eIF4B IκBα
Filamin A
eEF2K GSK-3 DAPK METTL1 BAD nNos
|
Erk1/2 p90RSK |
MKP-3 |
|
TSC2 |
|
|
|
Cell Adhesion |
PEA-15 |
Cytoplasmic Anchoring |
WAVE-2 |
|
|
PLD1 |
Ca+2-regulated |
|
|
|
|
Migration |
|
Exocytosis |
|
|
|
|
|
|
|||
Cytoplasm


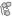











 Nucleus
Nucleus
MKP-1/2
cdc25 |
PPARγ |
|
|
|
|
Erk1/2 |
MSK1/2 |
p27 KIP1 |
p90RSK |
BUB1 |
|
|
|
|
|||
ER |
Ets |
Elk-1 |
TIF1A FoxO3 CREB |
ATF1 |
ER81 |
SRF |
c-Fos |
ERα |
Nur77 |
MITF |
C/EBPβ |
Stat1/3 |
c-Myc/ |
Pax6 |
c-Fos |
Histone |
HMGN1 |
ETV1 |
TIF1A |
ATF4 |
Myt1 |
Mad1 |
YB1 |
N-Myc |
|||||||||||
|
|
|
|
H3 |
|
|
|
|
|
|
|
UBF |
|
|
|
|
|
|
|
|
|
|
|
Transcription
Pathway Diagram Keys
Kinase |
Enzyme |
|
G-protein |
|
|
|
|
|
Direct Inhibitory |
|
Tentative Inhibitory |
|
|
Translocation |
||||||||
Phosphatase |
pro-apoptotic |
|
Acetylase |
|
Modification |
|
Modification |
|
|
|
Transcriptional |
|||||||||||
|
|
|
|
|
|
|||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||
|
|
Multistep Stimulatory |
|
|
|
|
Separation of Subunits |
|
|
Stimulatory Modification |
||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
Transcription Factor |
pro-survival |
|
Deacetylase |
|
Modification |
|
or Cleavage Products |
|
|
|
Transcriptional |
|||||||||||
|
|
|
|
|
|
|||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||
Caspase |
GAP/GEF |
|
|
|
|
|
|
|
|
|
Multistep Inhibitory |
|
|
|
Joining of Subunits |
|
|
Inhibitory Modification |
||||
|
Ribosomal subunit |
|
Modification |
|
|
|
|
|
|
|
||||||||||||
Receptor |
GTPase |
|
Direct Stimulatory |
|
Tentative Stimulatory |
|
|
|
|
|
|
|
||||||||||
|
|
|
|
|
|
|
|
|||||||||||||||
|
|
Modification |
|
Modification |
|
|
|
|
|
|
|
|||||||||||
|
|
|
|
|
|
|
|
|
|
|
||||||||||||
© 2003 – 2014 Cell Signaling Technology, Inc.
