
205
state by the presence of a homologous dominant suppressor gene (called a tumor suppressor gene). Consider what happens, when that suppressor is lost either due to mutation, deletion, or improper mitotic segregation so that a daughter cell receives two copies of the homologous chromosome with the recessive oncogene on it. These are all rare events, but once they
occur, the resulting cells have a growth advantage; there is a selection in their favor. In the case of a loss due to deletion, the resulting cells will be hemizygous in the region containing the oncogene. Strictly speaking, they will show only single alleles for all polymorphisms in
this region, and thus a zone of apparent homozygosity will mark the location of the oncogene. In the case of improper mitotic segregation, the entire chromosome carrying the onco-
gene may be homozygous, and once this is discovered, it is useful for diagnostic purposes but less useful for finding the oncogene, since an entire chromosome is still a very large target to search.
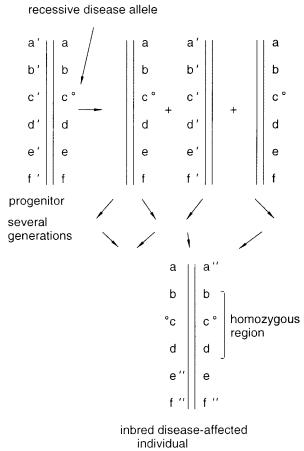
A second application of homozygous regions is the genetic technique known as homozygosity mapping. This is illustrated in Figure 6.38. It is useful in those relatively rare
Figure 6.38 Example of homozygosity mapping. Inbreeding within a family can reveal an ancestral disease allele by a pattern of homozygous alleles surrounding it.

206 GENETIC ANALYSIS
BOX 6.6
IMPRINTING
For a small number of genes in the human and mouse, the phenotype caused by an al-
lele depends |
on |
the parent |
who |
transmitted it. Some alleles are |
preferentially ex- |
pressed from |
the |
gene coded |
for |
by the father’s gamete (for |
example, insulinlike |
growth factor 2 and small ribonucleoprotein peptide n [Snrpn] in the mouse). Others are expressed preferentially from the mother’s gamete (such as insulinlike growth factor 2 receptor and H19 in the mouse). These effects can be seen strikingly when one parent or the other contains a deletion in an imprinted region.
DNA methylation is believed to play the major role in marking which parent of origin will dominate. Generally methylation silences gene expression, so methylation of a region in a particular parent will silence the effect of that parent. The process of imprinting is complicated. Imprinting may not be felt the same in all tissues in which the gene is expressed. The imprint must be established in the gametes; it may be maintained during embryogenesis and in somatic adult tissues, but must be erased in the early germline so that
a new imprint can be re-established in the gametes of the individual who is now ready to transmit it to the next generation. There are well-known examples of human diseases that show a parent of origin phenotype arising from differences in methylation. For example,
Prader-Willi syndrome is |
used to |
describe |
the |
disease phenotype when a damaged or |
deleted human analog of the |
mouse |
Snrpn gene |
is |
inherited from the father, whereas |
Angelman syndrome describes the rather different phenotype when the deleted allele is inherited from the mother. For further reading see Razin and Cedar (1994).
cases where there is significant inbreeding in the human population. Consider a progenitor carrying a recessive disease allele. Several generations later come two individuals who have inherited this allele from the original progenitor mate and have offspring together.
One or more offspring receive two copies of the disease allele, and as a result they are affected by the inherited disease. Because these offspring have inherited the allele from the same original chromosome, they are likely to share not only that allele but also nearby alleles. While the chromosome will have suffered several recombination events during the generations that have ensued between the progenitor and the affected individuals, most of
the original chromosome has presumably remained intact. This approach will be useful in
finding disease genes that segregate |
in |
inbred populations, and it has the advantage that |
only affected individuals need to |
be |
examined to find the zone of homozygosity that |
marks the approximate location of the disease gene.
A third application of homozygous regions is their frequent occurrence in the rather remarkable genetic phenomenon called imprinting. Here, as described in Box 6.6, it matters from which parent a gene is inherited.
SOURCES AND ADDITIONAL READINGS
Churchill, G. A., Giovannoni, |
J. J., and Tanksley, S. D. 1993 Pooled-samplings makes high-resolu- |
||
tion |
mapping practical with DNA markers. |
Proceedings of the National Academy of Sciences |
|
USA |
90: 16–20. |
|
|
|
|
|
|
SOURCES |
AND ADDITIONAL READINGS |
207 |
Dib, C., Fauré, S., Fizames, C., Samson, |
D., et al. 1996. A comprehensive genetic map of the hu- |
|
||||
man genome based on 5,264 microsatellites. |
|
Nature |
380: 152–154. |
|
||
Dietrich, W. F., Miller, J., Steen, R., Merchant, M. A., et al. 1996. A comprehensive genetic map of |
|
|||||
the mouse genome. |
Nature |
380: 149–152. |
|
|
|
|
Donis-Keller, H., Green, P., Helms, C., et al. 1987. A genetic linkage map of the human genome. |
|
|||||
Cell 51: 319–337. |
|
|
|
|
|
|
Lalouel, J.-M., and White, R. L. 1996. Analysis of genetic linkage. In Rimoin, D. L., Connor, J. M., |
|
|||||
and Pyeritz, R. E., eds. |
Principles |
and Practice |
of Medical Genetics |
. New York: Churchill |
||
Livingstone, pp. 111–125. |
|
|
|
|
|
|
Lander, E., and Botstein, D. 1986. Mapping complex genetic traits in humans: New methods using |
|
|||||
a complete RFLP linkage map. |
|
Cold Spring Harbor Symposium |
51: 1–15. |
|||
Lander, E., and Botstein, D. 1987. Homozygosity mapping: A way to map human recessive traits |
|
|||||
with the DNA of inbred children. |
|
|
Science |
236: 1567–1570. |
|
|
Leppart, M., Dobbs, M., Scambler, P. et al. 1987. The gene for familial polyposis coli maps to the |
|
|||||
long arm of chromosome 5. |
|
Science |
238: 1411–1413. |
|
|
|
Lindahl K. F. 1991. His and hers recombinational hotspots. |
|
Trends in Genetics |
7: 273–276. |
|||
Nicholls, R. D., Knoll, J. H. M., Butler, M. G., Karam, S., and Lalande, M. 1989. Genetic imprint-
ing suggested by maternal heterdisomy in non-delection Prader-Willi syndrome. Nature 342: 281-286.
Ott, J. 1991. |
Analysis |
of Human Genetic Linkage |
, rev. ed. Baltimore: John |
Hopkins |
University |
|
Press. |
|
|
|
|
|
|
Razin, A. and Cedar, H. 1994. DNA methylation and genomic imprinting. |
Cell |
77: 473–476. |
||||
Roeder, G. S. 1990. Chromosome synapsis and genetic recombination: their roles in meiotic chro- |
|
|
||||
mosome segregation. |
Trends in Genetics |
6: 385–389. |
|
|
||
Shuler, G. D., Boguski, M. S., Stewart, E. A., Stein, L. D., et al. 1996. A gene map of the human |
|
|
||||
genome. |
Science |
275: 540–546. |
|
|
|
|
Swain, J. L., Stewart, T. A., and Leber, P. 1987. Parental legacy determines methylation and ex- |
|
|
||||
pression of |
an |
autosomal |
transgene: A molecular mechanism |
for parental imprinting. |
|
Cell 50: |
719-727. |
|
|
|
|
|
|
