
- •Nucleotides as metabolic regulators
- •ATP is not quite what it seems
- •GTP-binding proteins, G proteins, or GTPases
- •G proteins
- •The GTPase cycle: a monostable switch
- •Switching off activity: switching on GTPase
- •The G protein receptor kinase family
- •Receptor mechanisms obviating G proteins
- •Monomeric GTP-binding proteins
- •Ras proteins discovered as oncogene products
- •Subfamilies of Ras
- •Structure
- •Post-translational modifications
- •GTPases everywhere!
- •Mutations of Ras that promote cancer
- •Functions of Ras
- •RasGAPs
- •RasGAP
- •Mechanism of GTPase activation
- •Guanine nucleotide exchange factors (GEFs)
- •Essay: Activation of G proteins without subunit unit dissociation
- •Pheromone-induced mating response in yeast
- •Monitoring subunit interactions in living cells by FRET
- •References

GTP-Binding Proteins and Signal Transduction
Table 4.4 Monomeric GTPase post-translational lipid modifications
GTPase |
Target sequence |
Modification |
|
|
|
H-Ras |
CVLS |
farnesyl and palmitoyl |
|
|
|
K-Ras |
CVIM |
farnesyl |
|
|
|
N-Ras |
CVVM |
farnesyl and palmitoyl |
|
|
|
Rho family (Rac and |
CxxL |
farnesyl, geranyl and |
Rho) |
|
palmitoyl |
|
|
|
Rab |
CC/CxC |
geranylgeranyl |
|
|
|
ARF |
N-terminal glycine |
myristoyl |
|
|
|
Monomeric GTP-binding proteins
Ras proteins discovered as oncogene products
The Ras proteins are often referred to as proto-oncogene products. This is because they were first discovered as the transforming products91,92 of a group of related retroviruses, including the Harvey murine (H) virus93 and Kirsten sarcoma (K) virus.94 The transforming genes are fusions of the viral gag gene (see page 324) and one of the ras genes derived from the rats through which the virus had been passaged.
N-Ras was discovered as a transforming gene product having sequence homology to the other Ras proteins present in a neuroblastoma cell line.95,96 They are all single-chain polypeptides, 189 amino acids in length, bound to the plasma membranes of cells by post-translational lipid attachments at their C-termini (Table 4.4). They all bind guanine nucleotides (GTP and GDP) and they are GTPases.97,98 Evidence for a link to human tumours came with the finding that cultured fibroblasts transfected with DNA derived from a human tumour cell line contain a mutated form of Ras.99
In a high proportion of human tumours one of the three endogenous cellular forms of Ras is altered by somatic mutations that inhibit the rate of GTP hydrolysis.100 This ensures that they are in a persistently activated state. On the other hand, non-oncogenic forms, c-Ras, are present in all cells. These are regulators of cell growth and differentiation (Chapter 12).
Subfamilies of Ras
The sequences of the Ras proteins are closely related (Figure 4.15). The first 164 amino acids of human H-Ras and chicken Ras differ in only 2 positions, and the sequences of the first 80 amino acids of human N-Ras and Drosophila D-Ras
Gag (glycosylated antigen) is the gene encoding the internal capsid of the viral particle. crk (C10 regulator of kinase).
The incidence of mutated Ras proteins varies among different types of tumour. 90% of human pancreatic adenocarcinomas
and 50% of colon adenocarcinomas are associated with Ras mutations. They are rarely found in adenosarcoma of the breast.
101

Signal Transduction
Fig 4.15 Main features of the Ras primary sequence.The mammalian Ras proteins (H, K, and N) share very close identity for the first 164 amino acids (green indicates identity, mauve indicates a conservative substitution). With the exception of the cysteine residue (186), the C-terminal segment is highly divergent (yellow). The lower sections of the figure illustrate the details of sequence motifs involved in binding to the guanine nucleotide, the effector domain, and the C-terminal Caax box that forms the substrate for post-translational modification by isoprenylation. The residues marked in red are associated with oncogenic mutations
Yeast RAS activates adenylyl cyclase.101 The proteins RAS1 and RAS2, having 321 and 309 residues, are longer than their mammalian counterparts, though the first 80 residues still maintain a high (80%) degree of homology. Yeast cells lacking both
RAS1 and RAS2 are nonviable, but viability can be restored if they are induced to express the homologous mammalian H-Ras.102,103 (By convention, capital letters are used to indicate yeast genes.)
are identical. These close similarities are supported by many conservative substitutions. As with the -subunits, it is likely that this high degree of conservation has been driven by the need for these proteins to communicate with a large number of other components, including the activators, inhibitors and effectors (see Figure 4.19).
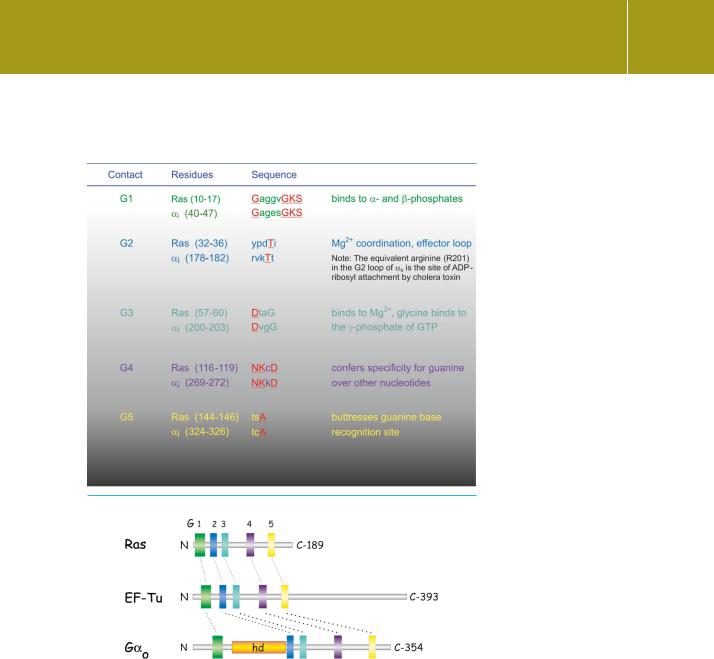
The Ras proteins are archetypes of a large superfamily. All members share some sequence homology to Ras and then fall into distinct groups, called Ras, Rho, Rab, Ran, Arf, and Kir/Rem/Rad (see Table 4.6). Within each subfamily, the homologies are rather strong. Beyond the more immediate subgroups of the Ras superfamily, all these proteins share some limited sequence homologies with short ‘fingerprint’ sequences present in the bacterial elongation factors (involved in protein synthesis) and also in the -subunits of the heterotrimeric G proteins (Table 4.5 and Figure 4.16). Indeed, the presence of these short motifs, appropriately distributed along the chain of a protein, can be taken as a fairly sure indication that it will be a GTPase. More than this, the presence
of -strands immediately adjacent to these highly conserved motifs is an invariant feature, whether they are Ras-related, elongation factors, or - subunits. Not too surprisingly, these conserved motifs constitute the sites of contact with guanine nucleotides.
Structure
A stereoscopic image of Ras is shown in Figure 4.17. Also shown are the elements of secondary structure and connecting regions, G1–G5, that form
102
GTP-Binding Proteins and Signal Transduction
Table 4.5 Conserved nucleotide binding motifs in H-Ras and bovine i. Binding contacts in red bold; non-conserved residues in lower case
Fig 4.16 Conserved nucleotide binding motifs in Ras, G , and EF-Tu.Three families of GTPases have generally divergent sequences but possess short stretches (G1–G5) that are similar to each other. When folded, these segments are almost superimposable and comprise the guanine nucleotide binding pocket. The motifs G2 and G3 (blue and cyan) lie within the switch regions 1 and 2 that undergo a conformational change when GDP is exchanged for GTP. hd indicates the -helical domain.From Bourne.6
the nucleotide binding pocket in colours corresponding to Table 4.5 and Figure 4.16. These motifs are also present in the elongation factors and the -subunits of heterotrimeric G proteins. Residues 26–45, encompassing the G2 contact, comprise the effector region that communicates with downstream proteins.
103

Signal Transduction
Fig 4.17 RasGDP and the motifs that form the nucleotide binding site. (a) RasGDP structure (stereo pair). (b) The conserved motifs G1–G5 that make contact with the nucleotide. They are depicted in colours that match those of Table 4.5. The bound nucleotide is GDP (spheres). The Mg2 ion is coloured green. Instructions for viewing stereo-images can be found on page xxv (4q2163–65,106).
Within this region, residues 30–40 are conserved in all forms of Ras, from yeast to mammals, and when mutated, the products are generally inactive (as measured in cell transformation assays: see page 306). Ras proteins mutated in the effector domain (switch region, see below) retain the ability to bind and catalyse the hydrolysis of GTP. Even when combined with a second (transforming) mutation that suppresses GTP hydrolysis and therefore prolongs the lifetime of the activated GTP bound state, the effector mutants
remain biologically inactive.104 The sequence of amino acids between residues 97 and 108 are responsible for interaction with guanine nucleotide exchange proteins (GEFs).105
Two regions of Ras, called the switch regions, change their conformation when the GDP is exchanged for GTP. These are indicated in Figure 4.18; here, the nucleotide binding site is occupied by the non-hydrolysable GTP analogue GppNHp. Both the switch I and switch II regions are implicated in the binding to effectors (such as the serine/threonine kinase Raf-1: see page 335) and to the GTPase-activating protein RasGAP. Switch I corresponds to the G2 loop that forms a part of the binding site for Mg2 and the switch II region includes G3 which makes contact with the -phosphate of GTP, and the following
104
