
228 |
CYTOGENETICS AND PSEUDOGENETICS |
|
|
|
|
|||
|
BOX 7.1 (Continued) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
QUANTITATIVE HIGH-RESOLUTION FISH |
|
|
|
|
|
|
|
|
mately linear function of the true distance, out to |
about 1.5 Mb (Fig. |
7.19 |
b ). Thus |
||||
|
these results support the use of a random walk model. The difficulty with the available |
|
|
|||||
|
results to date is that they do not agree on the scaling. Three studies are summarized |
|
|
|||||
|
below: |
|
|
|
|
|
|
|
|
|
INVESTIGATOR |
DISTANCES MEASURED |
REAL |
|
|
||
|
|
Lawrence |
|
1 |
|
1 Mb |
|
|
|
|
Trask |
|
1 |
|
0.5 Mb |
|
|
|
|
Skare |
|
1 |
|
1.2 Mb or more |
|
|
|
It is not clear if these considerable differences are due to |
differences in the methods |
|
|
||||
|
used to prepare the interphase chromatin or differences in the properties of interphase |
|
|
|||||
|
chromatin in different regions of the genome. Note that |
the scaling, at worst, is |
within |
|
|
|||
|
a factor of three of what was estimated in the text from a very crude model for the |
|
|
|||||
|
structure of the 30-nm filament. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
If this is taken to be a random walk, then the measured |
distance |
between markers should |
|
|
|||
|
be proportional to the square root of the real distance between them (Fig. 7.17 |
|
c;see |
also |
||||
|
Box 7.1). |
|
|
|
|
|
|
|
|
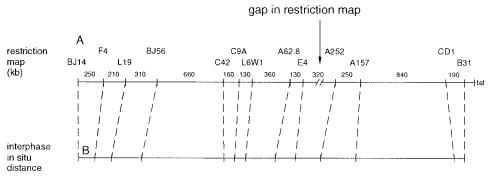
An example of the utility of interphase FISH is shown in Figure 7.20. Here interphase |
|
|
|||||
|
in situ hybridization was used to estimate the size of a gap that existed in the macrore- |
|
|
|||||
|
striction map of a region of human chromosome 4 near the tip of the short arm and now |
|
|
|||||
|
known to contain the gene for Huntington’s disease. |
The gap was present because no |
|
|
||||
|
clones or probes could be found between markers E4 and A252. In all other segments in |
|
|
|||||
|
the region, the accord between distances on the macrorestriction map and distances in- |
|
|
|||||
|
ferred from |
interphase in situ hybridization |
(using a |
scaling |
of 0.5 Mb per |
|
) is |
quite |
good. This has allowed the conclusion that the gap in the physical map would have to be small to maintain this consistency.
Figure 7.20 Comparison of a macrorestriction map of the tip of the short arm of human chromosome 4 with interphase FISH results for the same set of probes. (Adapted from Van den Engh et al., 1992.)
|
|
|
|
|
|
|
|
|
|
|
|
|
CHROMOSOME PAINTING |
229 |
|
CHROMOSOME |
PAINTING |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
For most chromosomes a dense set of mapped markers now exists. An even larger set of |
|
|
|
||||||||||||
clones is available and assigned to a particular chromosome but not yet mapped. These |
|
|
|
||||||||||||
clones can be used to examine the state of the entire chromosome, either in metaphase or |
|
|
|
||||||||||||
in less condensed states. This practice is called |
|
|
|
chromosome |
painting. |
Metaphase |
chro- |
||||||||
mosome painting is a useful tool to see |
if a chromosome is intact or |
has |
rearranged |
in |
|
|
|||||||||
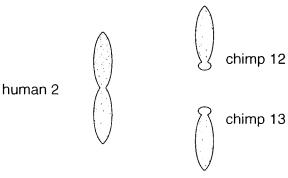
some way. A novel application of this is |
illustrated in Figure 7.21. Here |
the |
high |
degree |
|
|
|||||||||
of homology between chimp and human DNA sequences was used to examine the rela- |
|
|
|
||||||||||||
tionship between human chromosome 2 and its equivalent in the chimp. Probes from hu- |
|
|
|
||||||||||||
man chromosome 2 were used first on the human, where they indicated even and exhaus- |
|
|
|
||||||||||||
tive coverage of the chromosome. Next the same set of probes |
was |
used |
on |
the |
chimp. |
|
|
|
|||||||
Here two smaller chimp chromosomes were painted, numbers 12 and 13. Each of these is |
|
|
|||||||||||||
acrocentric, while human chromosome 2 is metacentric. These results make it clear that |
|
|
|||||||||||||
human chromosome |
2 |
must have |
arisen by |
a Robertsonian (centromeric) |
fusion |
of |
the |
|
|
|
|||||
two smaller chimp chromosomes. |
|
|
|
|
|
|
|
|
|
|
|
|
|||
When interphase chromatin is painted, one can observe the cellular location of particu- |
|
|
|||||||||||||
lar segments of DNA. The complexity of interphase chromatin makes it difficult to view |
|
|
|
||||||||||||
more than small DNA regions simultaneously. This technique is still in |
its infancy, |
but it |
|
|
|||||||||||
is already clear that it has the potential to provide an enormous amount of information of |
|
|
|||||||||||||
how DNA is organized in a functioning nucleus (Chandley et al., 1996, Seong et al., |
|
|
|||||||||||||
1994). |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
A recently developed variation of chromosome painting shows considerable promise |
|
|
|
||||||||||||
both in facilitating the search for gene involved in diseases like cancer and in assisting the |
|
|
|||||||||||||
development of improved clinical diagnostic tests for chromosomal disorders. In this |
|
|
|||||||||||||
technique, |
called |
|
comparative genome hybridization |
|
|
|
|
|
|
(CGH), total DNA from two sam- |
|
||||
ples to be compared is labeled using different specificity tags. Actual procedures use nick |
|
|
|||||||||||||
translation: For one sample a biotinylated dNTP is used; for the |
other |
a |
digoxigenin- |
|
|
||||||||||
labeled dNTP is used (see Chapter 3). These two samples are then allowed to hybridize |
|
|
|||||||||||||
simultaneously to |
a |
metaphase |
chromosome |
spread. The results |
are |
visualized by |
two- |
|
|
||||||
Figure 7.21 An example of metaphase chromosome painting, in which a human chromosome 2 probe is hybridized to the full set of human chromosomes, and separately to the full set of chimpanzee chromosomes. Shown are just those resulting chromosomes from both species that show significant hybridization.
230 CYTOGENETICS AND PSEUDOGENETICS
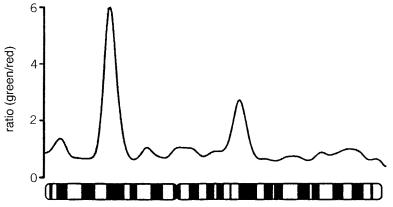
Figure 7.22 |
An example of comparative genome hybridization. Shown is the green to red color |
|
||
ratio seen along human chromosome 2 after competitive hybridization with DNA |
from two |
cell |
|
|
lines. The red cell line is normal human DNA. Three regions of DNA amplification are apparent in |
|
|||
the green labeled cell line. The most prominent of these corresponds to the oncogene |
N- |
myc |
known |
|
to be amplified in the cell line used. (From Kallioniemi et al., 1992.) |
|
|
|
|
color detection using fluorescein-labeled streptavidin or avidin to detect the biotin and rhodamine-labeled antidigoxigenin to detect the digoxigenin. The ratio of the two colors
should |
indicate the |
relative amounts of the two probes hybridized. This should be the |
same if |
the initial |
probes have equal concentrations everywhere in the genome. However, |
if there are regions in one target sample that are amplified or deleted, the observed color ratio will shift. The results can be dramatic as shown by the example in Figure 7.22. The color ratio shifts provide an indication of the relative amounts of each amplification or
deletion, |
and they also allow the |
locations of |
all |
such |
variations between |
two samples to |
be mapped in a single experiment. |
|
|
|
|
|
|
CHROMOSOME |
MICRODISSECTION |
|
|
|
|
|
Frequently |
the search for a gene |
has focused |
down |
to a |
small region of a |
chromosome. |
The immediate task at hand is to obtain additional DNA probes for this region. These are needed to improve the local genetic map and also to assist the construction of physical maps. They may also be useful for screening cDNA libraries if there is any hint of the preferred tissue for expression of the gene of interest. The question is how to focus on a small region of the genome efficiently. One approach has been microdissection of that region from metaphase chromosomes and microcloning of the DNA that results. Two approaches to microdissection are shown schematically in Figure 7.23. In one, a fine glass needle is used to scratch out the desired chromosome region and transfer the material to a
site |
where it can be collected. In the other |
approach, a laser is used |
to ablate all of the |
|||
genome except for the small chromosome region of interest. In either case, the problem in |
||||||
early |
versions |
of this method was to develop efficient |
cloning schemes |
that could start |
||
from |
the very |
small amounts of DNA that |
could |
be collected |
by |
microdissection. |

CHROMOSOME MICRODISSECTION |
231 |
Figure 7.23 |
Two |
examples of chromosome microdissection. ( |
a ) A fine needle is used to scratch a |
chromosome and |
transfer a |
section of it to another site where it can be further manipulated. ( |
b ) A |
laser is used to destroy the DNA in all but a preselected segment of a chromosome. |
|
||
Hundreds |
of |
microdissection |
products would be combined and placed in liquid micro- |
|||||||||||||||||
drops suspended in oil (Fig. 7.24). Restriction enzymes, vectors, and other components |
||||||||||||||||||||
needed for cloning were delivered |
by |
micromanipulators. |
The result |
was |
a |
technical |
||||||||||||||
tour |
de |
force |
|
that did deliver clones from the desired region but usually in very small |
||||||||||||||||
numbers. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
With |
the |
development |
of genome-wide PCR amplification methods (Chapter 4), |
||||||||||||||||
the |
need |
for |
microcloning |
dissected |
chromosome |
samples |
is |
eliminated. |
Using |
these |
||||||||||
methods, |
it |
is |
relatively |
easy to amplify the dissected |
chromosome |
material |
by |
PCR |
||||||||||||
until there |
|
is |
enough |
DNA |
to |
|
be |
handled |
by |
more |
conventional |
methods. |
Note |
|||||||
that the microdissection method has |
several important |
potential |
uses |
beyond |
the hunt |
|||||||||||||||
for |
specific |
disease genes. |
Most |
DNA |
libraries in |
any |
type |
of vector are |
biased, and |
|||||||||||
some regions are under-represented or not represented at all. Microdissection offers an attractive way to compensate for cloning biases, especially when they are severe. The alternate approach, in common use today, is to combine clones from different types of libraries in the hope that biases will compensate for each other. This can be a very effective strategy, but it increases substantially the number of DNA samples that must be handled. Another potential use for microdissection will be to pull clones from a particular individual who may have a region of special interest, such as a suspected rearrangement. This
will not always be an effective strategy, but it may well be necessary in cases where simpler approaches fail to yield a definitive picture of what has happened.
Figure 7.24 Microcloning of DNA in a tiny droplet containing microdissected chromosomes.
232 CYTOGENETICS AND PSEUDOGENETICS
SOURCES AND ADDITIONAL READINGS
Boyle, A. L., Feltquite, D. M., Dracopoli, N. C., Housman, D. E., and Ward, D. C. 1992. Rapid |
|
|
||||||||||
physical mapping of cloned DNA on banded mouse chromosomes by fluorescence |
|
|
|
|
in situ hy- |
|||||||
bridization. |
Genomics |
12: 106–115. |
|
|
|
|
|
|
||||
Burmeister, M., Kim, S., Price, E. R., de Lange, T., Tantravahi, U., Meyers, R. M., and Cox, D. |
|
|||||||||||
1991. A map of the distal region of the long arm of human chromosome 21 constructed by radi- |
|
|
||||||||||
ation hybrid mapping and pulsed field gel electrophoresis. |
|
|
Genomics |
9: 19–30. |
|
|||||||
Chandley, A. C., Speed, R. M., and Leitch, A. R. 1996. Different distributions of homologous chro- |
|
|
||||||||||
mosomes |
in |
adult human Sertoli cells and in |
lymphocytes signify nuclear differentiation. |
|
|
|||||||
Journal of Cell Science |
109: 773–776. |
|
|
|
|
|
|
|||||
Cherif, D., Julier, C., Delattre, O., Derré, J., Lathrop, G. M., and Berger, R. 1990. Simultaneous lo- |
|
|||||||||||
calization of cosmids and chromosome R-banding by fluorescence microscopy: Application to |
|
|
|
|||||||||
regional mapping of human chromosome 11. |
|
Proceedings of |
the National Academy |
of Sciences |
|
|||||||
USA |
|
87: 6639–6643. |
|
|
|
|
|
|
|
|||
Cox, D. R., Burmeister, M., Price, E. R., Kim, S., and Meyers, R. 1990. Radiation hybrid mapping: |
|
|
||||||||||
A somatic cell genetic method for constructing high-resolution maps of mammalian cell chro- |
|
|
||||||||||
mosomes. |
|
Science |
250: 245–251. |
|
|
|
|
|
|
|
||
Ellis, N., and Goodfellow, P. N. 1989. The mammalian pseudoautosomal region. |
|
|
|
Trends in Genetics |
||||||||
5: 406–410. |
|
|
|
|
|
|
|
|
|
|||
Heiskanen M., Kallioniemi, O., and Palotie, A. 1996. Fiber-FISH: Experiences and a refined proto- |
|
|||||||||||
col. |
Genetic Analysis (Biomolecular Engineering) |
|
12: 179–184. |
|
|
|||||||
Jeffreys, A. J., Neumann, R., and Wilson, V. 1990. Repeat unit sequence variation in minisatellites: |
|
|
||||||||||
A novel source of DNA polymorphism for studying variation and mutation by single molecule |
|
|
|
|||||||||
analysis. |
Cell |
60: 473–485. |
|
|
|
|
|
|
|
|||
Kallioniemi, A., Kallioniemi, O. P., Sudar, D., Rutovitz, D., Gray, J. W., Waldman, |
F., and Pinkel, |
|
|
|||||||||
D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. |
|
|
|
|||||||||
Science |
|
258: 818–821. |
|
|
|
|
|
|
|
|||
Lawrence, J., Singer, R. H., and McNeil, J. A. 1990. Interphase and metaphase resolution of differ- |
|
|
||||||||||
ent distances within the human dystrophin gene. |
|
Science |
249: 928–932. |
|
||||||||
Li, H., Cui, A., and Arnheim, N. 1990. Direct electrophoresis detection of the allelic state of single |
|
|
||||||||||
DNA molecules in human sperm by using the polymerase chain reaction. |
|
|
|
Proceedings of the |
||||||||
National Academy of Sciences USA |
87: 4580–4584. |
|
|
|
|
|
||||||
Li, H., Gyllensten, U. B., Cui, X., Saiki, R. K., Erlich, H. A., and Arnheim, N. 1988. Amplification |
|
|||||||||||
and analysis of DNA sequences in single human sperm and diploid cells. |
|
|
|
Nature |
335: 414–417. |
|||||||
Lichter, P., Ledbetter, S. A., Ledbetter, D. H., and Ward, D. C. 1990. Fluorescence in situ hybridiza- |
|
|
||||||||||
tion |
with |
Alu |
and |
L1 polymerase chain |
reaction probes |
for rapid |
characterization |
of human |
|
|
||
chromosomes |
in hybrid |
cell |
lines. |
Proceedings |
of the National Academy of |
Sciences USA |
87: |
|||||
6634–6638. |
|
|
|
|
|
|
|
|
|
|||
Lichter, P., Tang, C. C., Call, K., Hermanson, G., Evans, G. A., Housman, D., and Ward, D. C. |
|
|
||||||||||
1990. High resolution mapping of human chromosome 11 by in situ hybridization with cosmid |
|
|
|
|||||||||
clones. |
Science |
247: 64–69. |
|
|
|
|
|
|
|
|||
McCarthy, L. C. 1996. Whole genome radiation hybrid mapping. |
|
|
|
Trends in |
Genetics |
12: 491–493. |
||||||
Nilsson, M., Krejci, K., Koch, J., Kwiatkowski, M., Gustavsson, P., and Landegren, U. 1997. |
|
|||||||||||
Padlock probes reveal single nucleotide differences, parent of origin and in situ distribution of |
|
|
||||||||||
centromeric sequences in human chromosomes 13 and 21. |
|
|
Nature Genetics |
16: 252–255. |
||||||||
Ruano, G., Kidd, K. K., and Stephens, J. C. 1990. Haplotype of multiple polymorphisms resolved |
|
|
||||||||||
by enzymatic amplification of single DNA molecules. |
|
Proceedings |
of the National Academy |
of |
||||||||
Sciences USA |
87: 6296–6300. |
|
|
|
|
|
|
|
||||
|
|
|
|
SOURCES AND ADDITIONAL READINGS |
|
233 |
|
Seong, D. C., Song, M. Y., Henske, E. P., Zimmerman, S. O., Champlin, R. E., Deisseroth, A. B., |
|
|
|||||
and Siciliano, M. J. 1994. Analysis of interphase cells for the Philadelphia translocation using |
|
|
|||||
painting probe made by Inter- |
Alu |
-polymerase chain reaction from a radiation hybrid. |
Blood |
83: |
|||
2268–2273. |
|
|
|
|
|
|
|
Silva, A. J., and White, R. 1988. Inheritance of allelic blueprints for methylation patterns. |
|
Cell |
54: |
||||
145–152. |
|
|
|
|
|
|
|
Stewart, E. A., McKusick, K. B., Aggarwal, A., et al. 1997. An STS-based radiation hybrid map of |
|
|
|||||
the human genome. |
|
Genome Research |
|
7: 422–433. |
|
|
|
Strong, S. J., Ohta, Y., Litman, G. W., and Amemiya, C. T. 1997. Marked improvement of PAC and |
|
|
|||||
BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of ge- |
|
|
|||||
nomic DNA. |
Nucleic Acids Research |
|
25: 3959–3961. |
|
|
|
|
Van de Engh, G., Sachs, R., and Trask, B. J. 1992. Estimating genomic distance from DNA se- |
|
|
|||||
quence location in cell nuclei by a random walk model. |
Science |
257: 1410–1412. |
|
|
|||
Wang, D., and Smith, C. L. 1994. Large-scale structure conservation along the entire long arm of |
|
|
|||||
human chromosome 21. |
|
Genomics |
|
20: 441–451. |
|
|
|
Wu, B-L., Milunsky, A., Nelson, D., Schmeckpeper. B, Porta, G., Schlessinger, D., and Skare, J. |
|
|
|||||
1993. High-resolution mapping of probes near the X-linked lymphoproliferative disease (XLP) |
|
|
|||||
locus. Genomics |
|
17: 163 – 170. |
|
|
|
|
|
Yu, J., Hartz, J., Xu, Y., Gemmill, R. M., Korenberg, J. R., Patterson, D., and Kao, F.-T. 1992. |
|
|
|||||
Isolation, characterization, and regional mapping of microclones from a human chromosome 21 |
|
|
|
||||
microdissection library. |
American Journal of Human Genetics |
51: 263–272. |
|
|
|||
