
Dictionary of DNA and Genome Technology
.pdf
avian erythroblastosis virus
acts only on growing cells. In this approach, a well-washed population of bacteria (that includes auxotrophs) is exposed to penicillin in a minimal medium; the prototrophs (which can grow) are killed by the penicillin. The remaining cells are washed and re-plated on a complete medium to recover any auxotrophs. It’s important to note that, if the auxotrophs had developed through an in vitro process of mutagenization, it is essential that the cells be allowed to grow for several generations in complete medium prior to exposure to penicillin; this is because newly mutated cells contain a full complement of (prototrophic) enzymes, and the auxotrophic phenotype develops only after several rounds of cell division – during which the prototrophic enzymes are ‘diluted out’. A further requirement in this method is that only a low concentration of cells be used; this is because auxotrophs should not be allowed to grow on nutrients released by lysed prototrophs – as this would render them susceptible to lysis by the penicillin. For this reason, STREPTOZOTOCIN may be used in place of penicillin.
Auxotrophs may also be isolated by REPLICA PLATING. avian erythroblastosis virus See ERB.
avirulence gene See GENE-FOR-GENE CONCEPT.
Axenfeld–Rieger syndrome An autosomal dominant disorder involving eye defects and certain systemic abnormalities. The syndrome has been associated with mutations in the PITX2 gene (which encodes a transcription factor). In some patients the disorder is reported to involve aberrant splicing of preRNA; it has been suggested that variability in the extent of the splicing fault may be reflected in the observed variability of phenotypic manifestations [BMC Med Genet (2006) 7:59].
5-aza-2′-deoxycytidine A cell-permeant agent used e.g. for
studying DEMETHYLATION.
[Example of use: BioTechniques (2006) 41(4):461–466.] azaserine (O-diazoacetyl-L-serine) An agent with antimicrobial and antitumor activity produced by Streptomyces sp (a Grampositive bacterium). Azaserine inhibits the activity of certain enzymes, including phosphoribosylformylglycinamidine synthetase – thus inhibiting biosynthesis of purines and, hence,
nucleotides.
(cf. DON; see also HADACIDIN.)
AZT See ZIDOVUDINE.
18

B
B A specific indicator of ambiguity in the recognition site of a RESTRICTION ENDONUCLEASE or in another nucleic acid seqence; ‘B’ indicates C or G or T in DNA, C or G or U in RNA.
B-DNA The form of DNA that appears to be the most common type found in vivo (and on which is based the Watson–Crick model, the so-called ‘double helix’): a right-handed doublehelix of ~10.4 base pairs/turn and a diameter of ~20Å.
(See also RIGHT-HANDED HELIX.)
Bac-to-Bac® A BACULOVIRUS EXPRESSION SYSTEM, marketed
by Invitrogen (Carlsbad CA), in which a recombinant baculovirus genome – containing the gene of interest – is generated within specialized cells of Escherichia coli by site-specific transposition.
The strain of E. coli used in this system (DH10Bac™) contains a BACMID into which the gene of interest is inserted (to form an expression bacmid ).
Initially, the gene of interest is inserted into the polylinker of a small (4.8 kb) pFastBac™ plasmid. Within this vector, the inserted gene is positioned downstream of the (strong) promoter of the baculovirus polyhedrin gene; the expression cassette within the pFastBac vector is flanked by the terminal sequences, Tn7L and Tn7R, of transposon Tn7. The vector, containing the gene of interest, is then transfected into cells of the DH10Bac strain of E. coli, within which the expression cassette is transferred from the vector to the bacmid by sitespecific transposition.
The recombinant bacmid, containing the gene of interest, is isolated from E. coli and is then transfected into insect cells for the production of extracellular recombinant baculovirus.
The Bac-to-Bac system has been used e.g. in studies on the capsid protein of the beak-and-feather-disease virus (BFDV) [J Virol (2006) 80(14):7219–7225] and for studying the conformational states of a SARS virus protein [J Virol (2006) 80 (14):6794–6800].
(See also PFASTBAC DUAL VECTOR KIT.)
Bacillus anthracis A species of Gram-positive, spore-forming, typically square-ended rod-shaped bacteria, virulent strains of which form anthrax toxin (encoded by plasmid pXO1) and a poly-D-glutamic acid capsule (encoded by plasmid pXO2). B. anthracis grows on nutient agar; in biochemical tests it gives results similar to those from Bacillus cereus. B. anthracis is susceptible to the γ phage (gamma phage).
The Sterne strain of B. anthracis forms anthrax toxin but lacks the capsule – lack of the capsule permitting elimination of the pathogen by phagocytosis.
Ames strain: see entry BACTERIA (table).
Disease similar to anthrax has been reported to be caused e.g. by strains of Bacillus cereus [see PLoS Pathogens (2006) 2(11):e122].
(See also SORTASE.)
back mutation (reverse mutation) (1) Following a given (primary) mutation: a mutation that restores the original nucleotide sequence.
(2) An intragenic SUPPRESSOR MUTATION.
bacmid A replicon, first constructed in the 1990s, which can replicate as a plasmid in Escherichia coli and can be used to transfect (susceptible) insect cells.
A bacmid consists of a recombinant baculovirus genome (see BACULOVIRIDAE) which includes a plasmid origin of replication and also a target (insertion) site for transposon Tn7 (attTn7).
If a bacmid is present in E. coli, a gene cassette – flanked by the two terminal parts of Tn7 – can be transposed into the attTn7 site in the bacmid by site-specific transposition, given that the functions for transposition are provided in trans from a helper plasmid in the bacterium.
bacteria A term used to refer to some or all organisms of the domain BACTERIA. Note that this term has a lower-case ‘b’; an upper-case ‘B’ is used for the domain Bacteria because this is the name of a taxon (a specific taxonomic category).
Bacteria One of the two domains of prokaryotic organisms – the other being ARCHAEA. Organisms in these two domains differ e.g. in their 16S rRNA, in the composition of their cellwall macromolecules, in the composition of their membrane lipids and also in flagellar structure; moreover, in general, gene expression in the Archaea (in terms of transcription and translation) appears to resemble the eukaryotic mode of expression more closely than the bacterial mode.
The typical bacterial genome is a circular chromosome (i.e. ccc dsDNA). Cells may contain a single chromosome or may contain more than one copy per cell (the number depending on species and on growth conditions).
In some species of bacteria the genome is linear dsDNA: see e.g. BORRELIA BURGDORFERI. (A linear chromosome is also present e.g. in a species with one of the largest genomes sequenced thus far in bacteria (~9.7 Mbp): Rhodococcus sp RHA1 [Proc Natl Acad Sci USA (2006) 103:15582–15587].)
In general, only one type of chromosome is found in the cells of a given species. In Vibrio cholerae each cell contains two types of chromosome – one (chromosome I) being larger than the other (chromosome II); jointly, these two chromosomes form the genome of V. cholerae.
Bacterial chromosomes are extensively folded into compact bodies. (See also NUCLEOID.)
The size of the genome varies among different species. In a number of cases the genome has been completely sequenced. The table lists the approximate size of the genome in various species and includes sources of information on sequences.
In addition to chromosome(s), many bacteria contain one or more types of PLASMID; however, plasmids are not normally present in certain bacteria (e.g. Brucella and Rickettsia). In Rhodococcus sp RHA1 the large (~9.7 Mbp) genome consists of a linear chromosome and three linear plasmids.
Bacteria are commonly susceptible to one or more types of BACTERIOPHAGE, and a lysogenic strain of bacteria contains the phage genome (the prophage) in addition to the cell’s
19
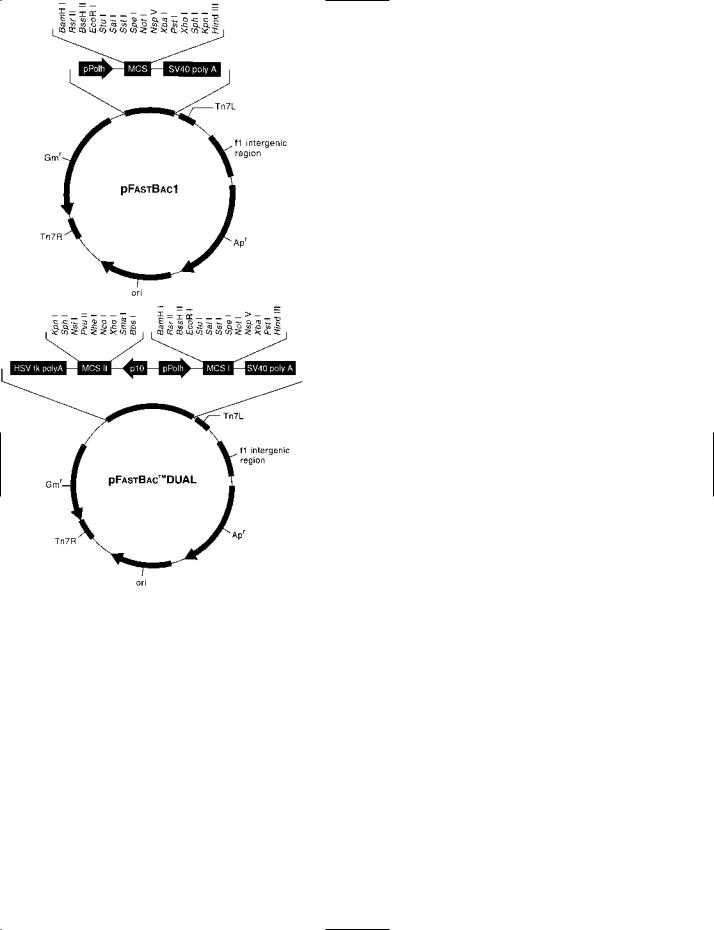
Bac-to-Bac® VECTORS Above. The basic pFastBac™1 vector. See entry for details of the method. Below. The pFastBac™ Dual vector into which two DNA sequences can be inserted and expressed simultaneously.
Courtesy of Invitrogen, Carlsbad CA, USA.
20

BACTERIAL GENOMES: examples of size and sources of information
Species |
Genome |
Reference |
|
(~Mbp)a |
|
Gram-positive |
|
Bacillus anthracis (Ames strain) |
5.2 |
Bacillus subtilis |
4.2 |
Bacteroides thetaiotaomicron |
6.3 |
Clostridium perfringens |
3.0 |
Lactobacillus acidophilus NCFM |
2.0 |
Lactobacillus plantarum |
3.3 |
Listeria welshimeri |
2.8 |
Mycobacterium avium subsp paratuberculosis |
4.8 |
Mycobacterium leprae |
3.3 |
Mycobacterium tuberculosis |
4.4 |
Rhodococcus sp RHA1 |
9.7 |
Streptococcus pneumoniae |
2.1 |
Streptococcus pyogenes |
1.9 |
Tropheryma whipplei |
0.9 |
Gram-negative |
|
Aeromonas hydrophila |
4.7 |
Borrelia burgdorferi |
0.9 |
Caulobacter crescentus |
4.0 |
Chlamydia trachomatis |
1.0 |
Coxiella burnetii |
2.0 |
Escherichia coli |
5.0c |
Escherichia coli O157:H7 |
5.5 |
Helicobacter pylori |
1.6 |
Helicobacter pylori (HPAG1) |
1.6 |
Neisseria meningitidis |
2.2 |
Pasteurella multocida |
2.2 |
Pseudomonas syringae pv. tomato |
6.5 |
Rhizobium leguminosarum |
7.7 |
Rickettsia typhi |
1.1 |
Shigella flexneri 5b |
4.5 |
Thiomicrospira crunogena |
2.4 |
Vibrio cholerae |
4.0d |
Nature (2003) 423:81–86
Nature (1997) 390:249–256
Science (2003) 299:2074–2076
PNASb (2002) 99(2):996–1001
PNASb (2005) 102(11):3906–3912
PNASb (2003) 100:1900–1995
J Bacteriol (2006) 188:7405–7415
PNASb (2005) 102(35):12344–12349
Nature (2001) 409:1007–1011
Nature (1998) 393:537–544
PNASb (2006) 103:15582–15587
Science (2001) 293:498–506
PNASb (2001) 98:4656–4663
Lancet (2003) 361:637–644
J Bacteriol (2006) 188(3):8272–8282 Nature (1997) 390:580–586
PNASb (2001) 98(7):4136–4141 Science (1998) 282:754–759 PNASb (2003) 100(9):5455–5460 Science (1997) 277:1453–1474
Nature (2001) 409:529–533; erratum (2001) 410:240 Nature (1997) 388:539–547
PNASb (2006) 103(26):9999–10004 Nature (2000) 404:502–506 PNASb (2001) 98:3460–3465
PNASb (2003) 100(18):10181–10186 Genome Biol (2006) 7(4):R34
J Bacteriol (2004) 186(17):5842–5855 BMC Genomics (2006) 7:62
PLoS Biol (2006) 4(12):e383
Nature (2000) 406:477–483
aApproximate genome size in Mbp (bp × 106). Variation in genome size can occur within a given species. bProc Natl Acad Sci USA.
cValues of ~4.7–5.5 have been reported for different strains.
dVibrio cholerae chromosome I ~3.0 Mbp, chromosome II ~1.0 Mbp (genome size: ~4.0 Mbp).
own genome. In some cases the prophage confers important characteristics on the host cell – e.g. the CTXΦ prophage (which encodes cholera toxin) is the major virulence factor in cholera-causing strains of Vibrio cholerae. Commonly, the prophage integrates in the bacterial chromosome; this is not the case, however, in e.g. phage P1.
Bacteria are used frequently in recombinant technology for a variety of reasons which include: (i) typically, rapid growth and division, (ii) ease of manipulation and mutagenesis, (iii) ease of storage, and (iv) (compared with eukaryotic cells)
cellular processes which are relatively easy to manage. In particular, Escherichia coli has been extensively used as an experimental organism. (Other bacteria that have been widely used as experimental organisms include e.g. Bacillus subtilis (Gram-positive), Pseudomonas aeruginosa (Gram-negative), and Salmonella typhimurium (Gram-negative).) Moreover, some bacterial components (e.g. plasmids and RESTRICTION ENDONUCLEASES) are invaluable in this area of study.
Before the 1980s, bacterial taxonomy was based on various phenotypic characteristics (including e.g. reaction to certain
21

bacterial two-hybrid system
dyes). Modern bacterial taxonomy is based primarily on the comparison of nucleic acids. Most TYPING methods are also genotypic (an important exception being PHAGE TYPING).
bacterial two-hybrid system See e.g. BACTERIOMATCH TWO-
HYBRID SYSTEM.
bacteriocin Any of a variety of (typically) protein or peptide factors, produced by certain bacteria (including both Gramnegative and Gram-positive species), which kill or inhibit other bacteria – frequently closely related species; they range from high-molecular-weight proteins and short peptides to a simple modified amino acid (microcin A15). (The activity of some bacteriocins requires post-translational modification.)
Analogous agents are produced by members of the Archaea (e.g. the halocins of halobacteria) but their amino acid sequences are apparently not homologous to those of their bacterial counterparts.
Bacteriocins are of various distinct types: see e.g. COLICIN,
LANTIBIOTIC and MICROCIN.
Most bacteriocins are encoded by genes in conjugative or non-conjugative plasmids. Chromosomally encoded bacteriocins include some of those formed by lactic acid bacteria and e.g. ‘bacteriocin 28b’ (a colicin from Serratia marcescens).
Bacteriocins act in characteristic ways. Some (e.g. microcin B17) inhibit DNA gyrase; some (e.g. cloacin DF13) cleave 16S rRNA; and some form pores in the bacterial cytoplasmic membrane.
BacterioMatch® two-hybrid system A system (Stratagene, La Jolla CA) for detecting interaction between two proteins in a bacterial cell. The proteins (bait and target) correspond to the ‘bait’ and ‘prey’ proteins in the YEAST TWO-HYBRID SYSTEM (q.v. for basic information on two-hybrid systems). Each of these proteins is encoded in a separate expression vector – in which the protein-encoding sequence is part of a fusion gene; the two vectors, pBT and pTRG, are introduced into a bacterial cell and expressed in the cell. [Use (e.g.): Appl Environ Microbiol (2007) 73(4):1320–1331.]
The bait vector contains the gene of the bait protein fused to the gene of the cI protein of PHAGE LAMBDA. The (DNAbinding) fusion protein (which is expressed in the cell) has a region that can bind to a λ operator sequence located a short distance upstream of the promoter of a reporter gene.
In the target vector, the gene of the target protein is fused to a sequence encoding the N-terminal region of the (bacterial) RNA polymerase α-subunit; the fusion protein is expressed in the cell.
Within the (engineered) reporter cell, interaction between bait and target proteins results in the recruitment of a functional RNA polymerase to the (nearby) downstream promoter, thus initiating transcription of the reporter gene.
The reporter gene encodes resistance to ampicillin; this allows the positive selection of cells in which protein–protein interaction has occurred (because these cells are able to grow on ampicillin-containing media). Another reporter gene, lacZ (encoding β-galactosidase), is transcribed in series with the Ampr gene; it can be used to validate interaction between the
two proteins.
In any bacterial two-hybrid system (and in the yeast twohybrid system), one problem is self-activation – in which e.g. a bait protein may activate the reporter system without first interacting with the target protein – leading to a false-positive result. This problem was addressed, in one approach, by a counter-selective procedure designed for use specifically in bacteria; this is based on the URA3/5-FOA mechanism (see YEAST TWO-HYBRID SYSTEM) which is modified for use in
Escherichia coli. In E. coli, the pyrF gene is homologous to
URA3, and a ∆pyrF strain of E. coli (in which the pyrF gene is inactivated) can be complemented by an introduced copy of the gene URA3. In this counter-selective system, URA3 is introduced on a plasmid and is used as the reporter gene in a one-hybrid arrangement: transcription of URA3 depends on interaction between the DNA-binding domain, DBD, and a specific binding sequence upstream of the URA3 reporter. As the DBD in this counter-selective system is covalently bound to the α subunit of RNA polymerase, an interaction between DBD and the specific binding site can recruit a functional RNA polymerase and promote transcription of URA3 – with the formation of a suicide substrate in the presence of 5-FOA. Accordingly, cells in which this occurs fail to grow. [Method: BioTechniques (2006) 40(2):179–184.]
bacteriophage (phage) Any virus that infects bacteria; many (perhaps most, or all) bacteria are susceptible to one or more types of phage. (See entries under PHAGE for some specific examples.)
A given phage may specifically infect bacteria of only one species or strain, or it may have a wider host range. Certain phages are specific for conjugative donor cells (i.e. ‘male’ cells), in which the PILUS forms the site of initial attachment.
(See also FEMALE-SPECIfiC PHAGES.)
Phages are often said to be virulent or temperate. A virulent phage lyses (kills) the bacterial host cell. A temperate phage can form a more or less stable relationship with a bacterium; in most cases the phage genome (the prophage) integrates in the bacterial chromosome, but in some cases (e.g. phage P1) it remains a circular, extrachromosomal ‘plasmid’. (See also
PHAGE N15.)
Under certain conditions a temperate phage may be induced to enter the lytic cycle – in which case virions are formed and cell lysis occurs; thus, a prophage may retain the potential for virulence, and in a population of lysogenic bacteria (i.e. bacteria which are hosts to a temperate phage), spontaneous induction may occur in a small number of cells.
Phages of the family Inoviridae (e.g. phages f1, fd, M13 in enterobacteria; Pf1 in Pseudomonas; Xf in Xanthomonas; v6 in Vibrio) are long filamentous phages which, in most or all cases, infect only ‘male’ (conjugative donor) cells. Although these phages replicate (and form virions) within the host cells they do not cause lysis: the virions are extruded through the cell envelope of the (living) host cells.
Phages commonly consist of nucleic acid enclosed within a protein coat called a capsid. However, certain phages contain
22

BacterioMatch® VECTORS The ‘bait’ and ‘target’ vectors (pBT and pTRG, respectively) of the BacterioMatch® two-hybrid system. See entry for details of the method. lcI refers to the gene of the cI protein of bacteriophage lambda (l). MCS = multiple cloning site (polylinker); P = promoter; ColE1 = origin of replication of the ColE1 plasmid; RNAPa = a region encoding the N-terminal sequence of the a subunit of (bacterial) RNA polymerase; Tetr = resistance to the antibiotic tetracycline; Camr = resistance to the antibiotic chloramphenicol.
Courtesy of Stratagene, La Jolla CA, USA.
lipid, either within the virion (for example, phage PM2 in
Alteromonas espejiana) or as an outer envelope (e.g. phage ϕ6 in Pseudomonas syringae p.v. phaseolicola), and certain phages contain components such as fucose or spermidine.
The phages vary greatly in shape and size. Many have a roughly spherical or icosahedral protein head (containing the genome) which carries a thin, elongated, contractile or noncontractile tail involved in attachment to the host cell. Some are small icosahedral virions which lack a tail, some have a complex, irregular shape, and some (as mentioned above) are long filaments.
In size, phages range from ~25 nm in diameter (e.g. ϕX174, Qβ – both tail-less, icosahedral phages) to ~200 nm (e.g. λ, T4 – both tailed phages), while filamentous phages (e.g. f1, M13) can be >750 nm in length, although they are only ~6 nm in width.
Genome
Depending on phage, the genome may be linear dsDNA (e.g. phages λ, P1, P2, P22, ϕ29, T4, T7); ccc dsDNA (e.g. PM2); ccc ssDNA (e.g. f1, ϕX174); ssRNA (e.g. MS2, Qβ) – or dsRNA (e.g. ϕ6).
Phage ϕ6 is unusual in that it has a SEGMENTED GENOME.
Phages in DNA technology
Phages, their components and/or the enzymes they encode are widely used in DNA technology (see the table for some examples).
Phages as potential therapeutic agents
Many studies have been carried out to investigate the use of phages against the bacteria that cause certain human diseases [see e.g. Antimicrob Agents Chemother (2001) 45:649–659]. Phage-encoded endolysins (enzymes that cleave the bacterial cell-wall polymer peptidoglycan) have also been considered
23

BACTERIOPHAGE: some uses of phages in DNA technology
Bacteriophage |
Genome |
Uses (e.g.) |
|
|
|
|
|
f1 |
ccc ssDNA |
• Replication origin used in pBluescript® phagemids |
|
|
|
• PHAGE DISPLAY technology |
|
λ (lambda) |
linear dsDNA |
• cI protein gene used in bacterial two-hybrid system (see BACTERIOMATCH TWO- |
|
|
|
|
HYBRID SYSTEM) |
|
|
• cos site used in cosmids and phasmids (see COSMID, PHASMID) |
|
|
|
• |
Derivatives used as cloning vectors (see e.g. ZAP EXPRESS VECTOR) |
|
|
• |
λ Red recombination system used for in vivo recombination (see the entry LAMBDA (λ) RED |
|
|
|
RECOMBINATION) |
|
|
• Constituents used in a range of GATEWAY SITE-SPECIfiC RECOMBINATION systems |
|
M13 |
ccc ssDNA |
• Used for making single-stranded copies of DNA target sequences for site-specific |
|
|
|
|
mutagenesis or sequencing etc. |
|
|
• Helper phage for phagemids, including e.g. gene-delivery phagemid particle vectors for |
|
|
|
|
eukaryotic cells (see entry GENE THERAPY) |
N15 |
linear dsDNA |
• Derivative used as a linear cloning vector in the BigEasy™ linear cloning system (q.v.) |
|
P1 |
linear dsDNA |
• Cre–loxP site-specific recombination system used e.g. in cassette-exchange mechanisms |
|
|
|
|
(e.g. RECOMBINASE-MEDIATED CASSETTE EXCHANGE) |
ϕ29 (phi 29) |
linear dsDNA |
• MULTIPLE DISPLACEMENT AMPLIfiCATION |
|
T3 |
linear dsDNA |
• RNA polymerase used for in vitro transcription |
|
|
|
• RNA polymerase used for incorporation of labeled nucleotides |
|
T4 |
linear dsDNA |
• DNA polymerase used for incorporation of digoxigenin-labeled nucleotides |
|
|
|
• DNA ligase used for closing nicks |
|
|
|
• UvsX recombinase used for the (isothermal) recombinase polymerase amplification of DNA |
|
|
|
|
(see entry RPA, sense 1) |
T7 |
linear dsDNA |
• As for T3 |
|
|
|
• Eberwine technique |
|
|
|
• DNA polymerase (Sequenase®), helicase and single-strand binding protein (gp2.5) used in |
|
circular helicase-dependent amplification of plasmids (see entry CHDA)
as therapeutic agents [Exp Biol Med (2006) 231:366–377]. In an entirely different approach, a number of studies have assessed phages, or phagemid particles, for gene delivery in the genetic modification of mammalian cells and, hence, their possible use in GENE THERAPY; in this context, an engineered helper phage has been developed to enable phagemid particles to incorporate a specific, helper-encoded peptide ligand in order to facilitate binding to target cells [BioTechniques
(2005) 39:493–497].
Phage delivery vehicles were also used for DNA vaccines [see e.g. Infect Immun (2006) 74(1):167–174].
bacteriophage conversion See LYSOGENY.
bacteroid A term used to refer to certain forms of bacteria that are distinct (e.g. morphologically) from related, normal wildtype cells – e.g. the bacterial nitrogen-fixing cells found in root nodules of leguminous plants.
BaculoDirect™ A BACULOVIRUS EXPRESSION SYSTEM (In-
vitrogen, Carlsbad CA) in which a gene/fragment of interest
is inserted into a modified, linearized baculovirus genome in a 1-hour in vitro step – following which the recombinant genome can be transfected directly into insect cells (such as Sf21 cells). After 72 hours, the supernatant (containing extracellular virions) can be used to infect a fresh batch of insect cells.
Insertion of a gene of interest into the baculovirus genome is carried out by a technique involving the GATEWAY SITE-
SPECIfiC RECOMBINATION SYSTEM: the baculovirus genome
includes a pair of attR sites into which the attL-flanked target DNA can be inserted by the activity of LR Clonase.
In the LR Clonase reaction – in which the target DNA is inserted – part of the linearized baculovirus genome, which contains a lacZ gene, is excised and forms a by-product. Subsequent staining of the baculovirus-infected cells to detect the absence of lacZ can be used to check that the cells do not contain non-recombinant baculovirus vectors.
In the recombinant baculovirus vector, the target DNA is
24

baculovirus expression systems
driven by a strong promoter (the promoter of the polyhedrin gene), and the vector encodes a V5-His tag (see also NICKEL- CHARGED AFfiNITY RESIN) to facilitate purification of the product.
One version of this vector contains a sequence encoding the MELITTIN peptide which is intended to promote secretion of the expressed protein.
The expression of heterologous proteins in insect cells can also be carried out in a baculovirus-independent manner by using simpler vector systems (including Gateway® and topotype systems) in Drosophila Schneider S2 cells: see DES.
Baculoviridae A family of enveloped viruses that infect a wide range of arthropods – including members of the Coleoptera (beetles), Diptera (flies), Hymenoptera (ants, bees etc.) and Lepidoptera (butterflies, moths).
The baculovirus genome consists of ccc dsDNA and ranges from ~80 kbp to ~200 kbp, according to virus.
In DNA technology, these viruses are exploited e.g. in the
BACULOVIRUS EXPRESSION SYSTEMS used for the expression
of recombinant proteins.
The baculoviruses include two major groups: the NUCLEAR
POLYHEDROSIS VIRUSES (NPVs) and GRANULOSIS VIRUSES
(GVs).
OBs, ODVs
Very late in the phase of infection, in the cell’s nucleus, the replicated viral nucleocapsids are enclosed within envelopes. They are subsequently embedded in a crystalline matrix that is composed of a virus-encoded protein, forming so-called occlusion bodies (OBs); the viruses occluded within these bodies are called occlusion-derived virions (ODVs).
The OBs of the nuclear polyhedrosis viruses (also referred to as polyhedra, owing to their shape) contain multiple enveloped particles, each of which contains either a single nucleocapsid or multiple nucleocapsids, according to virus. On the other hand, in granulosis viruses, each of the OBs contains a single enveloped particle containing a single nucleocapsid or, rarely, two nucleocapsids.
In the NPVs the crystalline matrix of the occlusion body is composed of the protein polyhedrin. In the granulosis viruses the matrix protein is granulin.
The OBs serve as disseminative units: when the host dies, and OBs are ingested by a new, susceptible host, infectious virions are released in the midgut of the new host and initiate infection.
BVs
Baculoviruses also form so-called budded virions (BVs). The BVs are formed when nucleocapsids are transported to the cell’s plasma membrane and then bud from the surface into the extracellular environment to produce enveloped virions (BVs). The BVs spread the infection to other cells in the host.
Baculovirus genes/gene products
Baculovirus genes/gene products referred to in the literature include e.g. ie-1 (encoding a transregulatory protein activator of transcription of both early and late genes); the lef genes (late expression factor genes, encoding e.g. a primase and its
accessory factor); gene gp64 (encoding a low-pH-dependent protein, GP64, involved in virion attachment to the host cell membrane); and p143 (a helicase).
The gene encoding very late expression factor 1 (VLF-1) is apparently present in the genomes of all baculoviruses. The product, VLF-1, is reported to have homology with members of the tyrosine recombinase family; however, a function unrelated to recombinase action (in which VLF-1 is involved in the production of normal capsid particles) has been reported for this product [J Virol (2006) 80(4):1724–1733].
Biological control
In addition to their uses in DNA technology, baculoviruses have been widely used for the biological control of various insect pests; they are regarded as particularly suitable for this purpose because of their apparent lack of relationship to the animal and plant viruses. The baculoviruses have been used e.g. to control the European pine sawfly (Neodiprion sertifer) and the European spruce sawfly (Gilpinia hercyniae) as well as the Small White Butterfly (Artogeia rapae).
baculovirus Any virus of the family BACULOVIRIDAE. baculovirus expression systems Systems in which vectors that
are based on the baculovirus genome (see BACULOVIRIDAE) are used for the expression of recombinant proteins in insect cells (or, more recently, in the cells of vertebrates). The use of baculovirus vectors for this purpose was suggested by the large quantities of matrix protein that are produced in insect cells infected with these viruses; as early as the 1980s, human interferon was produced in this kind of system.
Advantages of baculovirus expression systems
The advantages claimed for these systems include:
•High-level expression of recombinant proteins. Given the use of strong promoters to drive recombinant genes, yields of recombinant protein have been reported to reach up to ~500 mg/L (up to ~50% of total cell protein).
•Unlike prokaryotic cells, insect cells can carry out various post-translational modifications of expressed proteins similar to those carried out in mammalian cells.
•Procedures for growing (culturing) insect cells are simpler than those involved in the growth of mammalian cells – e.g. relatively simple, serum-free media can be used, and there is no requirement for CO2.
•High-density suspension culture can be employed.
•Protein expression can be easily scaled-up.
•Insect cells are characteristically refractory to infection by human viruses – so that recombinant proteins are unlikely to be contaminated by viral pathogens.
Baculovirus expression systems have been used e.g. for the production of veterinary products which include a vaccine against swine fever.
Disadvantages of baculovirus expression systems
These expression systems have several disadvantages:
•Some of the required post-translational modifications are poorly represented, or lacking, in insect cells. For example, patterns of glycosylation may be significantly different from those that characterize human glycoproteins. To combat this,
25

BaeI
genetically modified cells (see e.g. MIMIC SF9 INSECT CELLS) can be used; these cells are designed to express stably various mammalian glycosyltransferases.
• Recombinant proteins expressed in insect cells may remain largely intracellular. This is contrary to the general requirements of a production process – in which the formation of extracellular (i.e. secreted) protein facilitates the downstream processing of the product. In some systems, the gene of the recombinant protein is fused with a tag (e.g. a tag encoding multiple histidine residues – see NICKEL-CHARGED AFfiNITY RESIN) which can facilitate detection/isolation/purification of the product following cell lysis.
Development of baculovirus expression systems
Early procedures used for inserting a foreign gene into the baculovirus genome depended initially on homologous recombination – insect cells were co-transfected with (i) baculovirus DNA and (ii) the gene of interest, present in a separate vector and flanked by (non-essential) baculovirus DNA (e.g. sequences from the polyhedrin gene). If the gene of interest inserts into the polyhedrin gene (homologous recombination), the plaque morphology of the resulting recombinant baculovirus will be altered, thus allowing identification of recombinant viruses (and, hence, separation from the background of non-recombinant viruses). This approach, however, was found to yield only a low proportion (<1%) of recombinant viruses.
Improved recovery of recombinant viruses was achieved by using linearized viral genomes, and a further improvement resulted from the prior removal from the viral genome of an essential sequence, ORF 1629; in this case, a functional viral genome is produced only if recombination occurs with the experimental DNA – which includes ORF 1629 as well as the target sequence.
A rapid, commercial system produces recombinant baculovirus genomes by a 1-hour procedure involving in vitro sitespecific (att-mediated) recombination between a linearized, modified baculovirus genome and the cloned gene of interest:
see BACULODIRECT.
(See also BAC-TO-BAC and PFASTBAC DUAL VECTOR KIT.)
A convenient method for producing recombinant baculoviruses involves the (directional) insertion of target DNA via a simplified, low-cost technique which employs a restriction enzyme (Bsu36I) and the T4 DNA polymerase and T4 DNA ligase [see BioTechniques (2006) 41(4):453–458].
In some cases, expression of recombinant proteins has been achieved in whole (live) insects. For example, production of the veterinary product Vibragen Omega (used therapeutically for canine parvovirus infection) is carried out in silkworms; in this process, insects are infected with a recombinant polyhedrosis virus containing a sequence encoding recombinant feline interferon omega.
BaeI A type IIB RESTRICTION ENDONUCLEASE from Bacillus
sphaericus.
bait protein See YEAST TWO-HYBRID SYSTEM and BACTERIO- MATCH TWO-HYBRID SYSTEM.
BALB/c mice An inbred, laboratory strain of mice prone to the development of myelomas following certain stimuli.
barcoding See MOLECULAR INVERSION PROBE GENOTYPING.
Bardet–Biedl syndrome A genetically heterogeneous disorder involving a range of chromosomes (2, 3, 4, 7, 9, 11, 12, 14, 15, 16, 20) and exhibiting various phenotypic manifestations, including renal and retinal abnormalities.
barnase A bacterial endoribonuclease, produced by Bacillus amyloliquefaciens, which forms cyclic products.
Barr body (chromatin body; sex chromatin; X chromatin) A small, discrete body of CHROMATIN which can be seen in the nucleus in stained, interphase cells from female mammals. A Barr body is derived from an inactivated X CHROMOSOME (see X-INACTIVATION); hence, cells from a (normal) female contain one Barr body per nucleus, while cells from a normal male contain none.
In those individuals with atypical genotypes – such as XXX (superfemale) or XO (Turner’s syndrome) – the number of Barr bodies per nucleus is also atypical (respectively, two and zero in the superfemale and in individuals with Turner’s syndrome).
base composition See BASE RATIO.
base excision repair A DNA repair system, found in organisms ranging from bacteria to humans, that deals with chemically induced damage, including damage caused by reactive oxygen species.
The excision of an aberrant, chemically modified base is catalyzed by a DNA glycosylase. (See also 8-OXOG.) The phosphodiester bond on one side of the AP site is then cut by an AP endonuclease – e.g. exonuclease III in Escherichia coli (an enzyme which has exonuclease and apurinic/apyrimidinic endonuclease activity). Replacement of the damaged nucleotide (and apparently of several adjacent nucleotides) is then effected by a DNA polymerase.
(See also MISMATCH REPAIR.)
base J β-D-glucosylhydroxymethyluracil: an unusual base in the nuclear DNA of kinetoplastid protozoans and in Euglena.
In trypanosomatids, base J appears to replace a proportion of thymine residues and is found e.g. in the telomere region.
Attempts were made to obtain a null mutation in the gene of the base J-binding protein JBP1 but were not successful [Nucleic Acids Res (2005) 33(5):1699–1709].
base pairing (in double-stranded DNA and RNA) The pairing (i.e. binding, annealing, hybridization) of each base in one of the strands with a particular, complementary base in the other strand; such complementarity depends on the specificity of binding between cytosine and guanine, and between thymine (or uracil in RNA) and adenine.
base ratio (base composition; dissymmetry ratio) In DNA, the ratio:
[A+T]/[G+C]
in which A, T, G and C are relative molar amounts of the four types of nucleotide. Microbes with a base ratio of >1 are
26

binary deoxyribozyme ligase
sometimes called AT types; those with a base ratio <1 are GC types.
(cf. GC%.)
BCIP 5-Bromo-4-chloro-3-indolyl phosphate: a substrate for the enzyme alkaline phosphatase used in PROBE LABELING.
bcr-abl (BCR-ABL) See ABL.
bDNA assay Branched DNA assay: a method for quantitation of a specific DNA or RNA target sequence. Based on SIGNAL AMPLIfiCATION, it avoids a major problem associated with TARGET AMPLIfiCATION methods such as PCR, i.e. potential contamination of apparatus and environment with amplicons from previous assays.
Essentially, a bDNA assay involves: (i) a target-specific PROBE that immobilizes target sequences on a solid support, and (ii) a second type of probe which links each immobilized target to a synthetic, multi-branched molecule of DNA (socalled bDNA) – which, in turn, binds many enzyme-linked probes; using a dioxetane substrate, the enzyme generates an amplified chemiluminescent signal whose intensity is related to the number of target sequences in the sample.
bDNA kits (Quantiplex™; Bayer Diagnostics, Leverkusen) have been used e.g. for assaying viruses such as HIV (the human immunodeficiency virus) [J Clin Microbiol (2003) 41:3361–3367; J Clin Microbiol (2004) 42:3012–3016] and the simian immunodeficiency virus [J Virol (2004) 78:5324– 5337].
Beijing/W See W-BEIJING STRAIN.
BENA435 A cell-permeant DNA-staining fluorophore, with absorption and emission maxima at 435 nm and 484 nm respectively (when complexed with DNA), which is reported to be non-toxic and photoactivated in vivo. The compound is apparently an intercalating agent; fluorescence is reported to be stronger with dA/dT homopolymers than with dG/dC homopolymers.
It appears that BENA435 is not related to any other DNAstaining agent.
[BENA435: Nucleic Acids Res (2006) 34(5):e43.]
(See also DNA STAINING.)
Benefix® A recombinant form of blood-clotting factor IX used for the management of hemophilia B.
benzene-1,3,5-triacetic acid See BTA. O6-benzylguanine labels See GENE FUSION (uses).
benzylpenicillin Syn. Penicillin G (see also table in the entry
ANTIBIOTIC).
Bernard–Soulier syndrome (hemorrhagiparous thrombocytic dystrophy) A rare autosomal recessive condition characterized by a tendency for abnormal/excessive bleeding. Genetic lesions associated with this condition – on chromosomes 3, 17 and 22 – affect genes encoding subunits of a multi-subunit receptor for the von Willebrand factor.
[Bernard–Soulier syndrome: Orphanet J Rare Dis (2006) 1: 46.]
BEVS Baculovirus expression vector system(s) (see BACULO-
VIRUS EXPRESSION SYSTEMS).
bgl operon See CATABOLITE REPRESSION.
BglF See CATABOLITE REPRESSION.
BglG See CATABOLITE REPRESSION.
BHK cells Cells derived from baby hamster kidney.
BiFC BIMOLECULAR flUORESCENCE COMPLEMENTATION.
bifunctional intercalating agent |
See INTERCALATING AGENT. |
bifunctional vector Syn. SHUTTLE VECTOR. |
|
BigEasy™ linear cloning system |
A cloning system (Lucigen |
Corporation, Middleton WI) which involves a linear plasmid vector (pJAZZ-KA) derived from the linear dsDNA genome of coliphage N15. (See PHAGE N15.) The prophage of phage N15 does not integrate into the bacterial chromosome but replicates as an extrachromosomal ‘plasmid’. The mechanism of replication in N15 has features in common with replication in some other (linear genome) phages (e.g. ϕKO2) and with replication in the bacterium Borrelia burgdorferi (which also has a linear genome); see BORRELIA BURGDORFERI for more details.
Advantages of a linear vector system include the ability to clone sequences (e.g. inverted repeats, AT-rich regions) that may exhibit instability when cloned in circular, supercoiled plasmid vectors.
bimolecular fluorescence complementation (BiFC) A method used for the detection of protein–protein interaction in vivo; in this method, the two proteins of interest are each tagged with a (non-fluorescent) fragment of a fluorophore (such as YELLOW flUORESCENT PROTEIN) so that interaction between the two proteins in vivo may result in juxtaposition of the two parts of the fluorophore – with consequent production of a fluorescent signal on appropriate excitation.
In the procedure called extended BiFC (exBiFC), the pair of proteins whose interaction is being studied are each tagged with a constitutively fluorescent domain (a different fluorophore for each protein) in addition to the fragment of fluorophore used for the basic BiFC reaction. This enables the researcher to follow both (i) the expression of each protein and (ii) the localization of each of the proteins; it also permits normalization of the BiFC signal in relation to the levels of expression of the two proteins.
[exBiFC (e.g.): BioTechniques (2006) 41(6):688–692.] [BiFC fragments: BioTechniques (2006) 40(1):61–66.] BiFC for demonstrating protein interaction in Arabidopsis
[Plant Physiol (2007) 143(2):650–660] and Saccharomyces cerevisiae [Eukaryotic Cell (2007) 6(3):378–387].
binary deoxyribozyme ligase A type of engineered DEOXY- RIBOZYME which can be used to re-code sequence information [Nucleic Acids Res (2006) 34(8):2166–2172].
Based on the MAXIZYME principle, this ligase consists of two (ssDNA) ‘half-deoxyribozymes’ which can be activated to become a functional enzyme by the binding of a sequencespecific ‘bridging oligonucleotide’. In the active enzyme, the two ‘half-deoxyribozymes’ are base-paired in their central regions to form a (dsDNA) stem; at each end of this stem the pair of single strands – one from each ‘half-deoxyribozyme’ – can act as recognition sequences. At each end of the stem, i.e. at the junction between double-stranded and single-stranded
27
