
- •Preface and Acknowledgments
- •Contents
- •Contributors
- •1: Embryology for Urologists
- •Introduction
- •Renal Development
- •Pronephros
- •Mesonephros
- •Metanephros
- •Development of the Collecting System
- •Critical Steps in Further Development
- •Anomalies of the Kidney
- •Renal Agenesis
- •Renal Aplasia
- •Renal Hypoplasia
- •Renal Ectopia
- •Renal Fusion
- •Ureteral Development
- •Anomalies of Origin
- •Anomalies of Number
- •Incomplete Ureteral Duplication
- •Complete Ureteral Duplication
- •Ureteral Ectopia
- •Embryology of Ectopia
- •Clinical Correlation
- •Location of Ectopic Ureteral Orifices – Male (in Descending Order According to Incidence)
- •Symptoms
- •Ureteroceles
- •Congenital Ureteral Obstruction
- •Pipestem Ureter
- •Megaureter-Megacystis Syndrome
- •Prune Belly Syndrome
- •Vascular Ureteral Obstructions
- •Division of the Urogenital Sinus
- •Bladder Development
- •Urachal Anomalies
- •Cloacal Duct Anomalies
- •Other Bladder Anomalies
- •Bladder Diverticula
- •Bladder Extrophy
- •Gonadal Development
- •Testicular Differentiation
- •Ovarian Differentiation
- •Gonadal Anomalies
- •Genital Duct System
- •Disorders of Testicular Function
- •Female Ductal Development
- •Prostatic Urethral Valves
- •Gonadal Duct Anomalies
- •External Genital Development
- •Male External Genital Development
- •Female External Genital Development
- •Anomalies of the External Genitalia
- •References
- •2: Gross and Laparoscopic Anatomy of the Upper Urinary Tract and Retroperitoneum
- •Overview
- •The Kidneys
- •The Renal Vasculature
- •The Renal Collecting System
- •The Ureters
- •Retroperitoneal Lymphatics
- •Retroperitoneal Nerves
- •The Adrenal Glands
- •References
- •3: Gross and Laparoscopic Anatomy of the Lower Urinary Tract and Pelvis
- •Introduction
- •Female Pelvis
- •Male Pelvis
- •Pelvic Floor
- •Urinary Bladder
- •Urethra
- •Male Urethra
- •Female Urethra
- •Sphincter Mechanisms
- •The Bladder Neck Component
- •The Urethral Wall Component
- •The External Urethral Sphincter
- •Summary
- •References
- •4: Anatomy of the Male Reproductive System
- •Testis and Scrotum
- •Spermatogenesis
- •Hormonal Regulation of Spermatogenesis
- •Genetic Regulation of Spermatogenesis
- •Epididymis and Ductus Deferens
- •Accessory Sex Glands
- •Prostate
- •Seminal Vesicles
- •Bulbourethral Glands
- •Penis
- •Erection and Ejaculation
- •References
- •5: Imaging of the Upper Tracts
- •Anatomy of the Upper Tracts and Introduction to Imaging Modalities
- •Introduction
- •Renal Upper Tract Basic Anatomy
- •Modalities Used for Imaging the Upper Tracts
- •Ultrasound
- •Radiation Issues
- •Contrast Issues
- •Renal and Upper Tract Tumors
- •Benign Renal Tumors
- •Transitional Cell Carcinoma
- •Renal Mass Biopsy
- •Renal Stone Disease
- •Ultrasound
- •Plain Radiographs and IVU
- •Renal Cystic Disease
- •Benign Renal Cysts
- •Hereditary Renal Cystic Disease
- •Complex Renal Cysts
- •Renal Trauma
- •References
- •Introduction
- •Pathophysiology
- •Susceptibility and Resistance
- •Epidemiological Breakpoints
- •Clinical Breakpoints
- •Pharmacodynamic Parameters
- •Pharmacokinetic Parameters
- •Fosfomycin
- •Nitrofurantoin
- •Pivmecillinam
- •b-Lactam-Antibiotics
- •Penicillins
- •Cephalosporins
- •Carbapenems
- •Aminoglycosides
- •Fluoroquinolones
- •Trimethoprim, Cotrimoxazole
- •Glycopeptides
- •Linezolid
- •Conclusion
- •References
- •7: An Overview of Renal Physiology
- •Introduction
- •Body Fluid Compartments
- •Regulation of Potassium Balance
- •Regulation of Acid–Base Balance
- •Diuretics
- •Suggested Reading
- •8: Ureteral Physiology and Pharmacology
- •Ureteral Anatomy
- •Modulation of Peristalsis
- •Ureteral Pharmacology
- •Conclusion
- •References
- •Introduction
- •Afferent Signaling Pathways
- •Efferent Signaling
- •Parasympathetic Nerves
- •Sympathetic Nerves
- •Vesico-Spinal-Vesical Micturition Reflex
- •Peripheral Targets
- •Afferent Signaling Mechanisms
- •Urothelium
- •Myocytes
- •Cholinergic Receptors
- •Muscarinic Receptors
- •Nicotinic Receptors
- •Adrenergic Receptors (ARs)
- •a-Adrenoceptors
- •b-Adrenoceptors
- •Transient Receptor Potential (TRP) Receptors
- •Phosphodiesterases (PDEs)
- •CNS Targets
- •Opioid Receptors
- •Serotonin (5-HT) Mechanisms
- •g-Amino Butyric Acid (GABA) Mechanisms
- •Gabapentin
- •Neurokinin and Neurokinin Receptors
- •Summary
- •References
- •10: Pharmacology of Sexual Function
- •Introduction
- •Sexual Desire/Arousal
- •Endocrinology
- •Steroids in the Male
- •Steroids in the Female
- •Neurohormones
- •Neurotransmitters
- •Dopamine
- •Serotonin
- •Pharmacological Strategies
- •CNS Drugs
- •Enzyme-inducing Antiepileptic Drugs
- •Erectile Function
- •Ejaculatory Function
- •Premature Ejaculation
- •Abnormal Ejaculation
- •Conclusions
- •References
- •Epidemiology
- •Calcium-Based Urolithiasis
- •Uric Acid Urolithiasis
- •Infectious Urolithiasis
- •Cystine-Based Urolithiasis
- •Aims
- •Who Deserves Metabolic Evaluation?
- •Metabolic Workup for Stone Producers
- •Medical History and Physical Examination
- •Stone Analysis
- •Serum Chemistry
- •Urine Evaluation
- •Urine Cultures
- •Urinalysis
- •Twenty-Four Hour Urine Collections
- •Radiologic Imaging
- •Medical Management
- •Conservative Management
- •Increased Fluid Intake
- •Citrus Juices
- •Dietary Restrictions
- •Restricted Oxalate Diet
- •Conservative Measures
- •Selective Medical Therapy
- •Absorptive Hypercalciuria
- •Thiazide
- •Orthophosphate
- •Renal Hypercalciuria
- •Primary Hyperparathyroidism
- •Hyperuricosuric Calcium Oxalate Nephrolithiasis
- •Enteric Hyperoxaluria
- •Hypocitraturic Calcium Oxalate Nephrolithiasis
- •Distal Renal Tubular Acidosis
- •Chronic Diarrheal States
- •Thiazide-Induced Hypocitraturia
- •Idiopathic Hypocitraturic Calcium Oxalate Nephrolithiasis
- •Hypomagnesiuric Calcium Nephrolithiasis
- •Gouty Diathesis
- •Cystinuria
- •Infection Lithiasis
- •Summary
- •References
- •12: Molecular Biology for Urologists
- •Introduction
- •Inherited Changes in Cancer Cells
- •VEGR and Cell Signaling
- •Targeting mTOR
- •Conclusion
- •References
- •13: Chemotherapeutic Agents for Urologic Oncology
- •Introduction
- •Bladder Cancer
- •Muscle Invasive Bladder Cancer
- •Metastatic Bladder Cancer
- •Conclusion
- •Prostate Cancer
- •Other Chemotherapeutic Drugs or Combinations for Treating HRPC
- •Conclusion
- •Renal Cell Carcinoma
- •Chemotherapy
- •Immunotherapy
- •Angiogenesis Inhibitor Drugs
- •Conclusion
- •Testicular Cancer
- •Stage I Seminoma
- •Stage I non-seminomatous Germ Cell Tumours (NSGCT)
- •Metastatic Germ Cell Tumours
- •Low-Volume Metastatic Disease (Stage II A/B)
- •Advanced Metastatic Disease
- •Salvage Chemotherapy for Relapsed or Refractory Disease
- •Conclusion
- •Penile Cancer
- •Side Effects of Chemotherapy
- •Conclusion
- •References
- •14: Tumor and Transplant Immunology
- •Antibodies
- •Cytotoxic and T-helper Cells
- •Immunosuppression
- •Induction Therapy
- •Maintenance Therapy
- •Rejection
- •Posttransplant Lymphoproliferative Disease
- •Summary
- •References
- •15: Pathophysiology of Renal Obstruction
- •Causes of Renal Obstruction
- •Effects on Prenatal Development
- •Prenatal Hydronephrosis
- •Spectrum of Renal Abnormalities
- •Renal Functional Changes
- •Renal Growth/Counterbalance
- •Vascular Changes
- •Inflammatory Mediators
- •Glomerular Development Changes
- •Mechanical Stretch of Renal Tubules
- •Unilateral Versus Bilateral
- •Limitations of Animal Models
- •Future Research
- •Issues in Patient Management
- •Diagnostic Imaging
- •Ultrasound
- •Intravenous Urography
- •Antegrade Urography and the Whitaker Test
- •Nuclear Renography
- •Computed Tomography
- •Magnetic Resonance Urography
- •Hypertension
- •Postobstructive Diuresis
- •References
- •Introduction
- •The Normal Lower Urinary Tract
- •Anatomy
- •Storage Function
- •Voiding Function
- •Neural Control
- •Symptoms
- •Flow Rate and Post-void Residual
- •Voiding Cystometry
- •Male
- •Female
- •Neurourology
- •Conclusions
- •References
- •17: Urologic Endocrinology
- •The Testis
- •Normal Androgen Metabolism
- •Epidemiological Aspects
- •Prostate
- •Brain
- •Muscle Mass and Adipose Tissue
- •Bones
- •Ematopoiesis
- •Metabolism
- •Cardiovascular System
- •Clinical Assessment
- •Biochemical Assessment
- •Treatment Modalities
- •Oral Preparations
- •Parenteral Preparations
- •Transdermal Preparations
- •Side Effects and Treatment Monitoring
- •Body Composition
- •Cognitive Decline
- •Bone Metabolism
- •The Kidneys
- •Endocrine Functions of the Kidney
- •Erythropoietin
- •Calcitriol
- •Renin
- •Paraneoplastic Syndromes
- •Hypercalcemia
- •Hypertension
- •Polycythemia
- •Other Endocrine Abnormalities
- •References
- •General Physiology
- •Prostate Innervation
- •Summary
- •References
- •Wound Healing
- •Inflammation
- •Proliferation
- •Remodeling
- •Principles of Plastic Surgery
- •Tissue Characteristics
- •Grafts
- •Flap
- •References
- •Lower Urinary Tract Symptoms
- •Storage Phase
- •Voiding Phase
- •Return to Storage Phase
- •Urodynamic Parameters
- •Urodynamic Techniques
- •Volume Voided Charts
- •Pad Testing
- •Typical Test Schedule
- •Uroflowmetry
- •Post Voiding Residual
- •Further Diagnostic Evaluation of Patients
- •Cystometry with or Without Video
- •Cystometry
- •Videocystometrography (Cystometry + Cystourethrography)
- •Cystometric Findings
- •Comment:
- •Measurements During the Storage Phase:
- •Measurements During the Voiding Phase:
- •Abnormal Function
- •Disorders of Sensation
- •Causes of Hypersensitive Bladder Sensation
- •Causes of Hyposensitive Bladder Sensation
- •Disorders of Detrusor Motor Function
- •Bladder Outflow Tract Dysfunction
- •Detrusor–Urethral Dyssynergia
- •Detrusor–Bladder Neck Dyssynergia
- •Detrusor–Sphincter Dyssynergia
- •Complex Urodynamic Investigation
- •Urethral Pressure Measurement
- •Technique
- •Neurophysiological Evaluation
- •Conclusion
- •References
- •Endoscopy
- •Cystourethroscopy
- •Ureteroscopy and Ureteropyeloscopy
- •Nephroscopy
- •Virtual Reality Simulators
- •Lasers
- •Clinical Application of Lasers
- •Condylomata Acuminata
- •Urolithiasis
- •Benign Prostatic Hyperplasia
- •Ureteral and Urethral Strictures
- •Conclusion
- •References
- •Introduction
- •The Prostatitis Syndromes
- •The Scope of the Problem
- •Category III CP/CPPS
- •The Goal of Treatment
- •Conservative Management
- •Drug Therapy
- •Antibiotics
- •Anti-inflammatories
- •Alpha blockers
- •Hormone Therapies
- •Phytotherapies
- •Analgesics, muscle relaxants and neuromodulators
- •Surgery
- •A Practical Management Plan
- •References
- •Orchitis
- •Definition and Etiology
- •Clinical Signs and Symptoms
- •Diagnostic Evaluation
- •Treatment of Infectious Orchitis
- •Epididymitis
- •Definition and Etiology
- •Clinical Signs and Symptoms
- •Diagnostic Evaluation of Epididymitis
- •Treatment of Acute Epididymitis
- •Treatment of Chronic Epididymitis
- •Treatment of Spermatic Cord Torsion
- •Fournier’s Gangrene
- •Definition and Etiology
- •Risk Factors
- •Clinical Signs and Symptoms
- •Diagnostic Evaluation
- •Treatment
- •References
- •Fungal Infections
- •Candidiasis
- •Aspergillosis
- •Cryptococcosis
- •Blastomycosis
- •Coccidioidomycosis
- •Histoplasmosis
- •Radiographic Findings
- •Treatment
- •Tuberculosis
- •Clinical Manifestations
- •Diagnosis
- •Treatment
- •Schistosomiasis
- •Clinical Manifestations
- •Diagnosis
- •Treatment
- •Filariasis
- •Clinical Manifestations
- •Diagnosis
- •Treatment
- •Onchocerciasis
- •References
- •25: Sexually Transmitted Infections
- •Introduction
- •STIs Associated with Genital Ulcers
- •Herpes Simplex Virus
- •Diagnosis
- •Treatment
- •Chancroid
- •Diagnosis
- •Treatment
- •Syphilis
- •Diagnosis
- •Treatment
- •Lymphogranuloma Venereum
- •Diagnosis
- •Treatment
- •Chlamydia
- •Diagnosis
- •Treatment
- •Gonorrhea
- •Diagnosis
- •Treatment
- •Trichomoniasis
- •Diagnosis
- •Treatment
- •Human Papilloma Virus
- •Diagnosis
- •Treatment
- •Scabies
- •Diagnosis
- •Treatment
- •References
- •26: Hematuria: Evaluation and Management
- •Introduction
- •Classification of Hematuria
- •Macroscopic Hematuria
- •Microscopic Hematuria
- •Dipstick Hematuria
- •Pseudohematuria
- •Factitious Hematuria
- •Menstruation
- •Aetiology
- •Malignancy
- •Urinary Calculi
- •Infection and Inflammation
- •Benign Prostatic Hyperplasia
- •Trauma
- •Drugs
- •Nephrological Causes
- •Assessment
- •History
- •Examination
- •Investigations
- •Dipstick Urinalysis
- •Cytology
- •Molecular Tests
- •Blood Tests
- •Flexible Cystoscopy
- •Upper Urinary Tract Evaluation
- •Renal USS
- •KUB Abdominal X-Ray
- •Intravenous Urography (IVU)
- •Computed Tomography (CT)
- •Retrograde Urogram Studies
- •Magnetic Resonance Imaging (MRI)
- •Additional Tests and Renal Biopsy
- •Intractable Hematuria
- •Loin Pain Hematuria Syndrome
- •References
- •27: Benign Prostatic Hyperplasia (BPH)
- •Historical Background
- •Pathophysiology
- •Patient Assessment
- •Treatment of BPH
- •Watchful Waiting
- •Drug Therapy
- •Interventional Therapies
- •Conclusions
- •References
- •28: Practical Guidelines for the Treatment of Erectile Dysfunction and Peyronie´s Disease
- •Erectile Dysfunction
- •Introduction
- •Diagnosis
- •Basic Evaluation
- •Cardiovascular System and Sexual Activity
- •Optional Tests
- •Treatment
- •Medical Treatment
- •Oral Agents
- •Phosphodiesterase Type 5 (PDE 5) Inhibitors
- •Nonresponders to PDE5 Inhibitors
- •Apomorphine SL
- •Yohimbine
- •Intracavernosal and Intraurethral Therapy
- •Intracavernosal Injection (ICI) Therapy
- •Intraurethral Therapy
- •Vacuum Constriction Devices
- •Surgical Therapy
- •Conclusion
- •Peyronie´s Disease (PD)
- •Introduction
- •Oral Drug Therapy
- •Intralesional Drug Therapy
- •Iontophoresis
- •Radiation Therapy
- •Surgical Therapy
- •References
- •29: Premature Ejaculation
- •Introduction
- •Epidemiology
- •Defining Premature Ejaculation
- •Voluntary Control
- •Sexual Satisfaction
- •Distress
- •Psychosexual Counseling
- •Pharmacological Treatment
- •On-Demand Treatment with Tramadol
- •Topical Anesthetics
- •Phosphodiesterase Inhibitors
- •Surgery
- •Conclusion
- •References
- •30: The Role of Interventional Management for Urinary Tract Calculi
- •Contraindications to ESWL
- •Complications of ESWL
- •PCNL Access
- •Instrumentation for PCNL
- •Nephrostomy Drains Post PCNL
- •Contraindications to PCNL
- •Complications of PCNL
- •Semirigid Ureteroscopy
- •Flexible Ureteroscopy
- •Electrohydraulic Lithotripsy (EHL)
- •Ultrasound
- •Ballistic Lithotripsy
- •Laser Lithotripsy
- •Ureteric Stents
- •Staghorn Calculi
- •Lower Pole Stones
- •Horseshoe Kidneys and Stones
- •Calyceal Diverticula Stones
- •Stones and PUJ Obstruction
- •Treatment of Ureteric Colic
- •Medical Expulsive Therapy (MET)
- •Intervention for Ureteric Stones
- •Stones in Pregnancy
- •Morbid Obesity
- •References
- •Anatomy and Function
- •Pathophysiology
- •Management
- •Optical Urethrotomy/Dilatation
- •Urethral Stents
- •Preoperative Assessment
- •Urethroplasty
- •Anastomotic Urethroplasty
- •Substitution Urethroplasty
- •Grafts Versus Flaps
- •Oral Mucosal Grafts
- •Tissue Engineering
- •Graft Position
- •Conclusion
- •References
- •32: Urinary Incontinence
- •Epidemiology and Risk Factors
- •Pathophysiology
- •Urge Incontinence
- •Conservative Treatments
- •Pharmacotherapy
- •Invasive/ Surgical Therapies
- •Stress Urinary Incontinence
- •Male SUI Therapies
- •Female SUI Therapies
- •Mixed Urinary Incontinence
- •Conclusions
- •References
- •33: Neurogenic Bladder
- •Introduction
- •Examination and Diagnostic Tests
- •History and Physical Examination
- •Imaging
- •Urodynamics (UDS)
- •Evoked Potentials
- •Classifications
- •Somatic Pathways
- •Brain Lesions
- •Cerebrovascular Accident (CVA)
- •Parkinson’s Disease (PD)
- •Multiple Sclerosis
- •Huntington’s Disease
- •Dementias
- •Normal Pressure Hydrocephalus (NPH)
- •Tumors
- •Psychiatric Disorders
- •Spinal Lesions and Pathology
- •Intervertebral Disk Prolapse
- •Spinal Cord Injury (SCI)
- •Transverse Myelitis
- •Peripheral Neuropathies
- •Metabolic Neuropathies
- •Pelvic Surgery
- •Treatment
- •Summary
- •References
- •34: Pelvic Prolapse
- •Introduction
- •Epidemiology
- •Anatomy and Pathophysiology
- •Evaluation and Diagnosis
- •Outcome Measures
- •Imaging
- •Urodynamics
- •Indications for Management
- •Biosynthetics
- •Surgical Management
- •Anterior Compartment Repair
- •Uterine/Apical Prolapse
- •Enterocele Repair
- •Conclusion
- •References
- •35: Urinary Tract Fistula
- •Introduction
- •Urogynecologic Fistula
- •Vesicovaginal Fistula
- •Etiology and Risk Factors
- •Clinical Factors
- •Evaluation and Diagnosis
- •Pelvic Examination
- •Cystoscopy
- •Imaging
- •Treatment
- •Conservative Management
- •Surgical Management
- •Urethrovaginal Fistula
- •Etiology and Presentation
- •Diagnosis and Management
- •Ureterovaginal Fistula
- •Etiology and Presentation
- •Diagnosis and Management
- •Vesicouterine Fistula
- •Etiology and Presentation
- •Diagnosis and Management
- •Uro-Enteric Fistula
- •Vesicoenteric Fistula
- •Pyeloenteric Fistula
- •Urethrorectal Fistula
- •References
- •36: Urologic Trauma
- •Introduction
- •Kidney
- •Expectant Management
- •Endovascular Therapy
- •Operative Intervention
- •Operative Management: Follow-up
- •Reno-Vascular Injuries
- •Pediatric Renal Injuries
- •Adrenal
- •Ureter
- •Diagnosis
- •Treatment
- •Delayed Diagnosis
- •Bladder and Posterior Urethra
- •Bladder Injuries: Initial Management
- •Bladder Injuries: Formal Repair
- •Anterior Urethral Trauma
- •Fractured Penis
- •Penile Amputation
- •Scrotal and Testicular Trauma
- •Imaging
- •CT-IVP (CT with Delayed Images)
- •Technique
- •Cystogram
- •Technique
- •Retrograde Urethrogram (RUG)
- •Technique
- •Retrograde Pyelogram (RPG)
- •Technique
- •One-Shot IVP
- •Technique
- •References
- •37: Bladder Cancer
- •Who Should Be Investigated?
- •Epidemiology
- •Risk Factors
- •Role of Screening
- •Signs and Symptoms
- •Imaging
- •Cystoscopy
- •Urine Tests
- •PDD-Assisted TUR
- •Pathology
- •NMIBC and Risk Groups
- •Intravesical Chemotherapy
- •Intravesical Immunotherapy
- •Immediate Cystectomy and CIS
- •Radical Cystectomy with Pelvic Lymph Node Dissection
- •sexual function-preserving techniques
- •Bladder-Preservation Treatments
- •Neoadjuvant Chemotherapy
- •Adjuvant Chemotherapy
- •Preoperative Radiotherapy
- •Follow-up After TUR in NMIBC
- •References
- •38: Prostate Cancer
- •Introduction
- •Epidemiology
- •Race
- •Geographic Variation
- •Risk Factors and Prevention
- •Family History
- •Diet and Lifestyle
- •Prevention
- •Screening and Diagnosis
- •Current Screening Recommendations
- •Biopsy
- •Pathology
- •Prognosis
- •Treatment of Prostate Cancer
- •Treatment for Localized Prostate Cancer (T1, T2)
- •Radical Prostatectomy
- •EBRT
- •IMRT
- •Brachytherapy
- •Treatment for Locally Advanced Prostate Cancer (T3, T4)
- •EBRT with ADT
- •Radical Prostatectomy
- •Androgen-Deprivation Therapy
- •Summary
- •References
- •39: The Management of Testis Cancer
- •Presentation and Diagnosis
- •Serum Tumor Markers
- •Primary Surgery
- •Testis Preserving Surgery
- •Risk Stratification
- •Surveillance Versus Primary RPLND
- •Primary RPLND
- •Adjuvant Treatment for High Risk
- •Clinical Stage 1 Seminoma
- •Risk-Stratified Adjuvant Treatment
- •Adjuvant Radiotherapy
- •Adjuvant Low Dose Chemotherapy
- •Primary Combination Chemotherapy
- •Late Toxicity
- •Salvage Strategies
- •Conclusion
- •References
- •Index

12
Molecular Biology for Urologists
Peter E. Clark
Introduction
The last several decades have seen an explosion of science and technology across all walks of life, and medicine is no exception. Improvements in engineering, optics, laser technology, pharmacology,and molecular biology have radically changed how we take care of patients on a daily basis. The development of the “Targeted Therapies” for advanced renal cell carcinoma (RCC) is just an one example of how discoveries in the basic biology of a disease have contributed to the development of novel therapeutics,thus markedly altering the standard of care for this disease. The field of urology has traditionally been at the forefront of these discoveries, as evidenced by the Nobel Prize winning work of Huggins and Hodges during the last century. As we move through the twenty-first century, a fundamental understanding of molecular biology will be increasingly important to understand the basis for the therapies we prescribe. The purpose of this chapter is to introduce the practicing urologist to the basics of molecular biology and its relevance to our field. It is impossible to relate all of molecular biology, in all its rich, intricate detail, in this short chapter. Therefore, the goal will be to illustrate some basic principles utilizing one of the most prominent examples relevant to urology today.The approach will be to introduce the basic principles of molecular biology using the biology of RCC as an example. The implications and how modern, targeted therapies fit into this biology will be emphasized.
The hope is that by learning the basic principles as they relate to one disease, the interested reader can go on to learn about the relevant pathways in other diseases as well using the same principles. Since it is impossible to give proper attention to every pathway and every disease, and the field changes so rapidly (indeed the pace of change is accelerating), the interested reader is encouraged to pursue specific interests in other pathways/disease via the many recent reviews cited throughout this chapter and elsewhere to explore these concepts in greater depth. Note that many of the terms introduced in this chapter (those that are in bold type and underlined) are listed in Table 12.1 with a brief definition.
Tumor Suppressor Genes
and VHL
One of the basic principles of molecular biology as it relates to the development of neoplasms (oncogenesis) is the concept of a tumor suppressor gene. These genes are important in regulating a variety of cellular processes, but their unifying theme is that when these genes’ (or their protein) function is lost in a cell, that cell becomes prone to malignant transformation. This fundamental concept grew out of the development of the tumor suppressor gene theory and the two-hit hypothesis as described by Knudson.1,2 Under this hypothesis, both copies of a tumor suppressor gene must be disabled in
C.R. Chapple and W.D. Steers (eds.), Practical Urology: Essential Principles and Practice, |
161 |
DOI: 10.1007/978-1-84882-034-0_12, © Springer-Verlag London Limited 2011 |
|

|
|
|
162 |
|
|
|
|
|
|
|
Practical Urology: EssEntial PrinciPlEs and PracticE |
Table 12.1. important definitions and concepts in molecular biology |
|||
Term |
Section |
Definition |
|
tumor suppressor gene |
a |
a gene encoding a protein which when lost tends to result in the formation of a |
|
|
|
|
neoplasm |
genome |
a |
all the genes found within a given organism |
|
somatic cell |
a |
the cells within a multicellular organism that do not produce gametes (oocyte |
|
|
|
|
or sperm).these represent the vast majority of cells in an organism |
germ line mutations |
a |
a permanent, heritable change in the dna sequence within the genome of a |
|
|
|
|
gamete (oocyte or sperm). as a result, it is present in every cell of any |
|
|
|
organism which develops from that cell |
chromosome |
a |
the structure within the cell nucleus that stores the dna in linear strands |
|
|
|
|
associated with a variety of proteins such as histones |
genetic changes |
B |
direct mutations or alterations in the dna sequence that are passed on from |
|
|
|
|
parent to progeny cells |
Epigenetic changes |
B |
Heritable changes in the expression of genes which do not involve a direct dna |
|
|
|
|
mutation. an example is dna methylation |
codon |
B |
a sequence of three adjacent nucleotides that codes for a particular amino acid. |
|
|
|
|
the sequence of these successive codons specifies the amino acid sequence |
|
|
|
of the resulting protein |
Frame shift mutation |
B |
a mutation (or change in dna) in which nucleotides are inserted or deleted |
|
|
|
|
such that the downstream sequence of codons is scrambled. since dna is |
|
|
|
read three nucleotides at a time (see codon above) a shift in the nucleotide |
|
|
|
sequence puts the entire remaining sequence out of frame, resulting in a |
|
|
|
scrambled and nonfunctional protein |
nonsense mutation |
B |
a mutation (or change in dna) in which an early or premature stop codon is |
|
|
|
|
created, resulting in a truncated protein that is typically nonfunctional. |
Missense mutation |
B |
a mutation (or change in dna) in which single nucleotide within a codon is |
|
|
|
|
changed resulting in a single amino acid change in a protein.this may or |
|
|
|
may not result in altered function |
dna methylation |
B |
Modification of dna in which a methyl group (cH3) is added to a cytosine that |
|
|
|
|
immediately precedes a quanine (cpg island).this is carried out by enzymes |
|
|
|
called dna methyl transferases (dnMt) |
Promoter |
B |
the site in the dna associated with a particular gene where the proteins |
|
|
|
|
responsible for transcription bind to initiate mrna expression. as a result, |
|
|
|
this site is where the expression of that particular gene is controlled |
response element |
c |
the dna nucleotide sequence in the promoter of a gene that is recognized by |
|
|
|
|
a regulatory protein that controls transcription, such as the HiFa/b |
|
|
|
heterodimer. Binding of the regulatory protein to its response element within |
|
|
|
a particular gene will either upor downregulate expression of that gene |
Messenger rna (mrna) |
c |
a single stranded rna molecule, coded for by the dna of a gene, which specifies |
|
|
|
|
the subsequent amino acid sequence of a protein during protein synthesis |
transcription |
c |
the process by which the genetic code in dna is transcribed into a complemen- |
|
|
|
|
tary, single stranded mrna molecule for subsequent protein synthesis |
translation |
c |
the process whereby the genetic code stored in the transcribed mrna molecule |
|
|
|
|
is now translated into a sequence of amino acids to form a protein.this is |
|
|
|
accomplished by the cooperative work of the ribosomes and trna that is |
|
|
|
associated with specific amino acids |

163
MolEcUlar Biology For Urologists
Table 12.1. (continued) |
|
|
Term |
Section |
Definition |
oncogene |
c |
a gene that is mutated or overexpressed in a cell, allowing it to become |
|
|
neoplastic or malignant |
Polymerase chain |
c |
a technique to significantly amplify a specific stretch of dna using a set of dna |
reaction (Pcr) |
|
primers designed to specify an area of interest |
rt-Pcr |
c |
a variation of Pcr in which mrna is reverse transcribed into dna and then Pcr |
|
|
is done on a gene of interest.this allows the scientist to measure gene |
|
|
expression at the mrna level |
genomics |
c |
a large-scale, rapid screen of gene expression across the entirety of the cell’s |
|
|
dna, that is, its genome |
Proteomics |
c |
a large-scale, rapid screen of the level of all the proteins within a cell at a given |
|
|
time |
angiogenesis |
d |
the process by which new blood vessels are formed |
ligand |
d |
a molecule, typically a soluble one such as a hormone, which binds to another |
|
|
molecule to initiate a subsequent downstream effect |
receptor |
d |
a molecule within or on the surface of a cell that when it binds its specific ligand |
|
|
(see above) it results in specific,downstream effects important to cellular function. |
Kinase |
d |
a frequently used abbreviation for a phosphokinase.these are enzymes which |
|
|
transfer a phosphate group from atP to another molecule |
cytoplasm |
d |
the portion of a eukaryotic cell that lies between the outer cellular surface |
|
|
membrane and the inner nucleus. it houses much of the cell’s physiologic |
|
|
machinery and organelles, such as the mitochondria, ribosomes, golgi |
|
|
apparatus, and endoplasmic reticulum |
nucleus |
d |
the large organelle that houses the genomic dna in eukaryotic cells within |
|
|
chromosomes |
Phosphorylation |
d |
the process whereby phosphate bonds are created between two substrates. in |
|
|
cells this is accomplished usually by kinases |
tyrosine kinase |
d |
an enzyme that transfers a phosphate group from atP to a tyrosine residue, |
|
|
either within its own structure or another molecule |
targeted therapy |
E |
a form of therapy that is designed to specifically target molecules important to |
|
|
the growth of cancer cells, rather than just target rapidly dividing cells such as |
|
|
cytotoxic therapy (i.e., traditional chemotherapy). Examples include |
|
|
antibodies to specific molecules and small molecule kinase inhibitors |
Monoclonal antibody |
E |
an antibody that can be produced in large quantities in a pure form from a |
|
|
single cell clone. Each antibody molecule is identical with respect to its |
|
|
structure and specificity for a particular target (termed an antigen) |
tyrosine kinase inhibitor |
F |
a small molecule that interferes with the tyrosine kinase activity of enzymes in |
|
|
cells.this is a prototypical example of a targeted therapy |
cell cycle |
g |
the regulated sequence by which cells undergo the process of growth and cell |
|
|
division/replication |
apoptosis |
|
also termed programmed cell death, it is the active, regulated process by which |
|
|
cells die in response to external or internal stimuli |
Heterodimer |
|
the association between two different proteins to form a complex. an example is the |
|
|
association between HiFaand HiFbin response to hypoxia or loss of VHl protein |
Homodimer |
|
the association between two identical proteins to form a complex. an example |
|
|
is the association between two androgen receptor molecules within a cell |

|
|
164 |
|
|
|
|
|
Practical Urology: EssEntial PrinciPlEs and PracticE |
order for a cancer to develop. In a sporadic, non- |
What followed was based on the recognition |
|
inherited form of cancer, this means that for any |
that these sporadic clear cell RCCs were also |
|
given cell in the body, a mutation is needed in |
seen in von Hippel-Lindau disease and the |
|
both copies of the same gene. This is because |
abnormality on chromosome 3p fit the profile of |
|
any mutation or change in the genome would be |
a putative tumor suppressor gene. In a series of |
|
in a somatic cell, and so, only that particular cell |
elegant and groundbreaking studies of multiple |
|
and its progeny would have that change, whereas |
different families with von Hippel-Lindau dis- |
|
the remaining cells in the body would not. It |
ease, the localization of the VHL gene was |
|
would be a rare event that each copy of a par- |
mapped to a relatively small region on chromo- |
|
ticular gene would acquire a mutation in the |
some 3p,18–20, which was then followed by the |
|
same cell; so in sporadic, nonfamilial cancers, |
identification of the VHL gene in a seminal arti- |
|
the tumors generally occur later in life and are |
cle by Latif et al. in 1993.21 Since that time, it has |
|
usually unifocal. On the other hand, in inherited |
been demonstrated that the majority of sporadic |
|
familial tumor syndromes, there is a germ line |
clear cell RCC tumors harbor aberrations of |
|
mutation in one tumor suppressor gene. Thus, |
VHL, strongly suggesting that the same gene |
|
every cell in the body has inherited a nonfunc- |
was responsible for both the inherited and non- |
|
tional copy of the gene (the first hit). As a |
inherited forms of the disease.1,2,22–26 Indeed, |
|
consequence, now it only takes one additional |
even in cases where mutations of VHL were not |
|
mutation of the same gene (the second hit) in |
identified, most often other aberrations were |
|
one cell in order for a cancer to develop. Since |
noted such as abnormal hypermethylation of |
|
every cell already has the first hit, having a sec- |
the promoter region of VHL, leading to low or |
|
ond hit would be more common. Patients suffer- |
absent protein levels (see below).27 |
|
ing from such inherited tumor syndromes |
Since the discovery of VHL, its role in the nor- |
|
would, therefore, develop cancers in the affected |
mal function of the cell and how it can act as a |
|
organs at a younger age and tend to have multi- |
tumor suppressor gene has been carefully stud- |
|
focal disease. The basic tenets of this hypothesis |
ied.The principles illustrated here are applicable |
|
were developed in the 1980s for familial tumor |
to a host of other disease processes and serve as |
|
syndromes such as retinoblastoma and the Rb |
an excellent example of how the discoveries at |
|
gene and neurofibromatosis and the NF-1 and |
the bench have been translated to the bedside in |
|
NF-2 genes.3–7 In urology, the classic example of |
the form of new targeted agents for advanced |
|
this is von Hippel-Lindau disease, the VHL gene, |
RCC. It is instructive, therefore, to describe this |
|
and the development of clear cell RCC. |
in some detail, though the interested reader is |
|
The discovery of the VHL gene and its rele- |
encouraged to read any one of the many in- |
|
vance to clear cell RCC grew at first from the |
depth reviews on this topic. |
|
observation that the RCC in von Hippel-Lindau |
|
|
disease was identical to its sporadic, nonfamilial |
Inherited Changes in Cancer Cells |
|
counterpart in every respect except it occurred |
||
earlier in life and tended to be multifocal. It |
|
|
therefore fit the profile of a familial tumor syn- |
It is worth pausing for a moment to review vari- |
|
drome characterized by the loss/mutation of a |
ous ways in which a gene, such as VHL, can |
|
tumor suppressor gene. Careful studies of three |
become aberrantly regulated. A critical element |
|
different families (kindreds) with a familial ten- |
for all of these mechanisms is that they can be |
|
dency toward the development of clear cell RCC |
inherited or passed on to any progeny cells. |
|
had consistently showed aberrations of the short |
Thus, once a “parent” or originating cancer cell |
|
arm of chromosome 3 (termed 3p).8–10 This |
acquires one of these changes, those changes are |
|
was followed by a series of studies of clear cell |
passed on to all its progeny resulting in the |
|
RCC tumors and cell lines that also showed |
growth of a tumor. The two fundamental forms |
|
abnormalities of chromosome 3p as a unifying |
of such changes are genetic changes, such as |
|
theme.11–15 The changes noted in chromosome 3 |
direct mutations of DNA,and epigenetic changes, |
|
were not present in normal tissues in these spo- |
inherited alterations in how genes are expressed |
|
radic clear cell RCC tumors and were not pres- |
(such as DNA methylation). Both of these mech- |
|
ent in other histologic RCC variants (such as |
anisms can be demonstrated in the VHL gene |
|
papillary RCC).16,17 |
and clear cell RCC. |
|

165
MolEcUlar Biology For Urologists
Genetic changes (mutations of the DNA in the |
them (de-acetylated via enzymes called histone |
genome) are inherited from one cell and passed |
de-acetylases or HDACs) or methyl groups |
on to all its progeny. If that cell is a gamete, such |
added (methylated via enzymes called histone |
as an egg or sperm, then this mutation will be |
methyltransferases, HMTs). In both cases, these |
found in every cell in the new organism. These |
can cause gene expression to be silenced.Perhaps |
are the types of mutations found in familial |
the most intensely studies form of epigenetic |
tumor syndromes such as von Hippel-Lindau |
change as it relates to cancer is the silencing of |
Disease. If a mutation occurs in a non-gamete |
gene expression via DNA methylation. This |
cell (i.e., any cell after conception), then this |
involves the covalent bonding of a methyl group |
somatic mutation is passed on only to that cell’s |
to a cytosine in areas where there is a cytosine |
progeny. Furthermore, such a somatic mutation |
immediately followed by a guanine, a sequence |
will not be passed on from generation to genera- |
known as a CpG island. This is carried out by a |
tion. These are the mutations found in sporadic |
class of enzymes called DNA methyltransferases |
cancers. The types of mutations that can occur in |
(DNMTs). When these CpG islands are methy- |
the DNA include insertions or deletions of small |
lated in the promoter of a gene, the gene’s expres- |
or large portions of the DNA. Since every three |
sion is often shut down. This mechanism is |
DNA nucleotides represents one codon (coding |
relevant to clear cell renal cell carcinoma, in that |
element for one amino acid), if the insertion or |
a significant fraction of sporadic RCCs have |
deletion occurs in a multiple of three, the overall |
been shown to lose VHL protein through DNA |
protein function may or may not be disrupted |
methylation and gene silencing, an epigenetic |
depending on what amino acid(s) is inserted/ |
change, rather than by a mutation.27 There is |
deleted. If the number of nucleotides inserted/ |
ongoing and intensive interest in the therapeutic |
deleted is not a multiple of three, then the entire |
potential of HDAC and DNMT inhibitors across |
sequence of nucleotide codons and resulting |
a variety of tumor types, though their direct |
amino acids will be altered resulting in a frame |
clinical utility in RCC remains to proven. |
shift mutation. This will, in effect, result in a |
|
scrambled series of amino acids and a nonfunc- |
The Tumor Suppressor VHL, |
tional protein. Roughly, half the VHL mutations |
|
found in sporadic clear RCC are of this type.22 |
Oncogene HIF, and Gene |
|
|
This will result in a completely nonfunctional |
Regulation |
protein. In some cases, the nucleotide may be |
|
mutated in a way that places a premature stop |
|
codon, so a portion of the protein is truncated. |
In a normal cell, the VHL protein acts predomi- |
These are referred to as nonsense mutations. In |
nantly to regulate the cell’s response to the local |
other cases, one nucleotide is mutated/changed |
availability of oxygen.33–38 When local oxygen |
such that only one amino acid is altered. This is |
levels are normal, there is another regulatory |
termed a missense mutation and represents the |
protein, termed hypoxia inducible factor alpha |
majority of the remaining VHL mutations in |
(HIFa), which is hydroxylated permitting it to |
sporadic, clear cell RCCs.22 A more in-depth |
bind to the VHL protein (see Fig. 12.1).39,40 The |
analysis of the specific VHL mutations and their |
VHL protein is actually part of an enzyme com- |
potential relationship to disease biology has |
plex (called an E3 ligase) that joins a series of |
been reviewed previously.28,29 |
molecules to HIFa called ubiquitin, which in |
The expression of genes can also be altered |
turn marks HIFa for degradation.41–46 In the |
via mechanisms that do not involve a mutation |
normal circumstance, then, HIFa levels are low |
in DNA. If these alterations are heritable, then |
in the cell. During hypoxia when oxygen levels |
these are termed epigenetic changes.30–32 The |
in the cell are low, HIFa is not hydroxylated, so |
ability to silence genes is critical to the normal |
it does not bind to VHL protein, and conse- |
development of an organism. There are a num- |
quently is not degraded. The levels of HIFa in |
ber of mechanisms by which this can be accom- |
the cell rise, allowing it to bind with a similar |
plished. This includes modifications of the |
molecule, HIF-b, that exists at high levels in the |
proteins that are involved in packaging DNA |
cell at all times (see Fig. 12.2). This complex can |
within the nucleus of cells called histones. These |
then bind to specific regions in the cell’s nuclear |
histones can have acetyl groups removed from |
DNA called hypoxia response elements (HRE). |
166
Practical Urology: EssEntial PrinciPlEs and PracticE
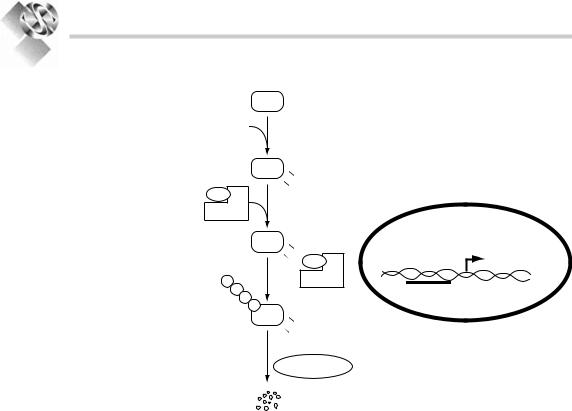
Figure 12.1. normal regulation of HiFa: under normal conditions, with normal levels of tissue oxygenation, HiFais hydroxylated through the action of enzymes termed prolylhyroxylases.this hydroxylation allows it to associate with an E3 ligase complex that includes the VHl protein.the VHl protein in particular is critical to this association.the E3 ligase then enzymatically tags HiFa with a series of molecules (called ubiquitin).this ubiquitination then marks HiFato be recognized and degraded by the proteasomal complex within the cell. HIFa Hypoxia inducible Factor alpha, VHL von Hippel lindau, Ub ubiquitin, HRE hypoxia response element.
|
HIFα |
p |
Cytoplasm |
|
p |
|
|
O2 |
Prolyl-hydroxylases |
|
|
|
|
||
|
HIFα |
p |
|
|
p |
OH |
|
VHL |
|
OH |
|
|
|
|
|
E3 ligase |
|
|
|
|
HIFα |
p |
|
|
p |
OH |
|
|
|
OH VHL |
|
Ub |
|
E3 ligase |
|
Ub |
|
|
HRE |
Ub |
|
||
|
|
||
|
Ub |
|
|
|
HIFα |
p |
|
|
p |
OH |
Nucleus |
|
|
OH |
|
|
|
|
|
Proteasome
These HREs are found in the regulator region, called the promoter, of genes important in the cell’s response to hypoxia. When a transcriptional regulator, such as the HIFa/b complex, binds to the response element (HRE) in the promoter of a gene, the transcriptional machinery of the cell is turned on and messenger RNA (mRNA) specific for that gene is produced in a process called gene transcription (see Fig. 12.3). These mRNA molecules then translocate to the cell’s cytoplasm where the code embedded in the mRNA is translated into a series of amino acids to generate the specific protein corresponding to that gene (gene translation). In the case of HIF and hypoxia, these correspond to a variety of genes that are critical to the cell’s response to hypoxia. These include vascular endothelial growth factor (VEGF), platelet derived growth factor (PDGF), transforming growth factor alpha (TGFa), carbonic anhydrase IX (CA-IX), erythropoietin, glucose transporter 1 (GLUT-1), and others. The VHLHIF axis serves as a good example, then, of a tumor suppressor (VHL) regulating an oncogene (HIFa) that in turn transcriptionally regulates a series of other genes that are important to normal cellular function.
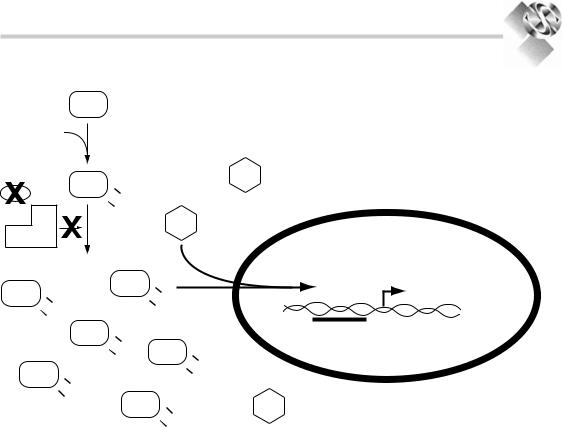
In the setting of clear cell RCC, the normal balance in the cell is upset. When the VHL protein either cannot function or is abnormally low/absent in the cell, then no matter the oxygen levels in the cell, HIFa is always at a high level since it cannot be marked for degradation (see Fig. 12.2). Thus,VHL protein is functioning as a tumor suppressor. High levels of HIFa in turn mean the HIFa/b complex will interact with the HREs in the nucleus and the genes normally regulated by HIF will be transcribed and translated at an abnormally high rate (see Fig. 12.3). It is this upregulation of genes such as VEGF, PDGF, and TGFa that is thought to lead to clear cell RCC. Therefore, HIFais acting as an oncogene.
It is worth pausing for a moment to review a number of general techniques and concepts that are important in the context of molecular biology. These concepts have been part of some of the studies discussed in this chapter and are an integral part of many, as yet unpublished work that is still ongoing in many centers across the world. These involve different ways of interrogating the genetic makeup of tissues (such as tumors) and how these tissues express different proteins that regulate their function.
167
MolEcUlar Biology For Urologists
HIFα p Cytoplasm
p
|
O2 |
|
|
|
|
|
Prolyl-hydroxylases |
|
|||
|
HIFα |
|
|
|
HIFβ |
|
|
|
|
|
|
|
|
OH |
|
|
|
|
|
OH |
|
|
|
|
|
|
|
HIFβ |
|
E3 ligase |
|
|
|
|
|
HIFα p |
|
HIFα |
|
|
|
|
|
|
OH |
|
|
|
|
|
|
|
|
p |
OH |
|
OH |
|
|
|
|
|
|
||
OH |
|
|
|
|
|
|
HIFα |
p |
|
|
HRE |
|
p |
OH |
HIFα |
p |
|
|
|
OH |
|||
|
|
|
p |
OH |
|
HIFα |
|
|
|
||
p |
|
|
|
OH |
|
|
|
|
|
|
|
|
p OH |
|
|
|
Nucleus |
|
OH |
HIFα |
|
|
|
|
|
p |
HIFβ |
||
|
|
|
|||
|
|
|
p |
OH |
|
|
|
|
|
OH |
|
Figure 12.2. accumulation of HiFa: under conditions of hypoxia, prolyl-hydroxylases cannot hydroxylate HiFa and so it will not bind to the VHl-E3 ligase complex. alternatively (as depicted here), if VHl protein is mutated, nonfunctional, or not present the VHl-E3 ligase cannot bind to HiFa. in either case, HiFais not ubiquitinated, and so is not degraded by the proteasome. it accumulates within the cell and associates with the constitutively present related molecule, HiFb. this heterodimer now translocates to the nucleus where it binds to specific regions
For example, one of the major advances in the study of biology has been the technique of the polymerase chain reaction (PCR). This is essentially a technique to significantly amplify a specific stretch of DNA using a set of DNA primers that the investigator can design to home in on an area of interest. The primers specify exactly what stretch of DNA is amplified. This technique has revolutionized the entire field of gene discovery and cloning. A variation of this can now be done whereby the mRNA in a cell can be reversely transcribed into DNA, and PCR then done on this, so called RT-PCR. This technique can now allow biologists to measure the relative expression levels of different genes in cells to see which are upor downregulated. Further refinements using these basic principles have
(hypoxia response elements or HrE’s) in the promoters of genes upregulated by HiF. the binding of the HiFa-HiFb heterodimer to this promoter region at the HrE turns on gene transcription. note that if VHl is absent or not functional, then no matter what the oxygenation level, HiFa cannot be recognized by the E3 ligase complex and is not degraded. HiFa accumulates in the cell even in the absence of hypoxia. HIFa Hypoxia inducible Factor alpha, HIFb Hypoxia inducible Factor beta, VHL von Hippel lindau, HRE hypoxia response element.
allowed such studies to be done on a mass scale using “gene chips” that now allow for measurement of thousands of genes at once. This large scale, rapid screen of gene expression across the entirety of the cell’s DNA,that is,its genome, is referred to as genomics. Other techniques can now screen the entirety of a cell’s protein mix using a variety of techniques generally referred to as proteomics. Indeed, the number of “–omics” has continued to expand including metabolomics, lipidomics, and others. Mass scale screenings of gene and protein expression is allowing investigators to create a signature for individual patients and tumors (or other diseased tissue). It is these approaches that hold the promise of “individualized” medicine for the future.
