
- •First steps in the isolation of steroid hormones
- •Origins
- •Beginning again
- •The discovery of intracellular hormone receptors
- •Evidence for intracellular receptors
- •A superfamily of nuclear receptors
- •Orphan receptors and evolution
- •Nomenclature of nuclear receptors
- •Receptor structure and ligand binding
- •Ligand-binding domains are molecular switches
- •Activation of cytosol-resident receptors
- •DNA binding
- •Recognizing response elements
- •Activation and repression of transcription
- •Coactivators
- •Corepressors
- •Transrepression
- •Regulatory networks
- •Interaction with other signalling pathways
- •Phosphorylation
- •Ligand-independent activation
- •Non-transcriptional actions of nuclear receptors and their ligands
- •References

Nuclear receptors
FIG 10.4 Domains of nuclear receptors.
Abbreviations: DBD, DNA-binding domain; LBD, ligand-binding domain; AF-1, activation function 1
(ligand-independent); AF-2, activation function 2 (ligand-dependent).
Nomenclature of nuclear receptors
Over the years, the study and classification of nuclear receptors has suffered from the problem of multiple names for the same gene. A systematic nomenclature has therefore been established,28 and official names are indicated in Figure 10.3. Full details are available from the bioinformatic database, Nurebase (http://www.ens-lyon.fr/LBMC/laudet/nurebase/ nurebase.html). More molecular specific information is provided by the nuclearRDB database.
Receptor structure and ligand binding
The general structure of the nuclear receptors is highly conserved, consisting of a set of common functional domains (see Figure 10.4). There is a highly variable N-terminal domain (A/B), a highly conserved DNA-binding domain (DBD) containing twinned zinc finger motifs, a hinge region (D) and a C- terminal segment (E/F) that contains the ligand-binding domain (LBD). There are two activation function sites, AF-1 and AF-2. AF-1 is in the N-terminal domain and is ligand-independent, while AF-2 is located in the LBD and is ligand-dependent. There are also sites that determine nuclear localization and dimerization.
Ligand-binding domains are molecular switches
The most common structure of the LBD is based on a compact assembly of 11 helices that form a three-layer sandwich (Figure 10.5). The middle layer does not extend all the way across, leaving a non-polar cavity at one end formed by the two outer layers. This is the binding pocket, and its size varies among the different receptors. For steroid and thyroid hormone receptors, VDR, RAR, and RXR, the cavity is matched to ligand size. These receptors bind their ligands with high affinity (KD in the nanomolar range). Then there is the group of receptors that bind with low affinity (micromolar range). These include LXR
281

Signal Transduction
FIG 10.5 Structure of the retinoic acid X receptor (RXR) without and with ligand.
Apo-RXR is shown on the left and the ligand-bound form on the right. Upon binding 9-cis retinoic acid (purple spheres), a conformational change occurs, involving mainly helix H12, which is amphipathic, closing the entrance to the pocket.
Rearrangements of H3 and the -loop also take place (1lbd29 and 1fby30).
and FXR, which have been termed metabolic sensors rather than receptors. They cope with a wider variety of ligands. The xenobiotic receptor PXR has one of the largest and most flexible cavities, with a volume of 1100 Å3, nearly twice that of the very high affinity TR receptor ( 600 Å3). Conversely, the orphan NURR1 has virtually no cavity.
The effect of ligand binding is a conformational change. In general, the molecule adopts a more compact structure with a rearrangement of the chains forming the cavity. This is particularly evident when RXR binds its ligand (Figure 10.5). The amphipathic C-terminal helix H12 (AF-2) swings across to close the pocket, trapping the ligand between the outer layers of the sandwich and a short two-stranded -sheet at the other end. For other receptors this movement is less distinct. In their unoccupied, apo forms, the region around the binding pocket (the lower end of the molecule in Figure 10.5) tends to be flexible (as a result, apo-receptors are difficult to crystallize). Binding of ligand then stabilizes the structure, which forms a more compact arrangement in which H12 becomes less mobile. The surface formed by H3 and H4 is altered so that receptor molecules bound at their DNA sites can interact with coregulator proteins (see below).
Activation of cytosol-resident receptors
Steroid receptors such as GR, which in the absence of a stimulus exist principally in the cytosol, cannot bind ligands until their ligand binding domains have been assembled in a receptive conformation. This is illustrated in Figure 10.6. Competence to bind hormone is achieved by the attachment of a series of heat shock proteins, including hsp70, hsp40, and most importantly hsp90, followed by a number of other accessory molecules. These include the regulatory cochaperone p23 and an immunophilin (e.g. FKBP52) which links the complex to the microtubule motor protein dynein. However,
282
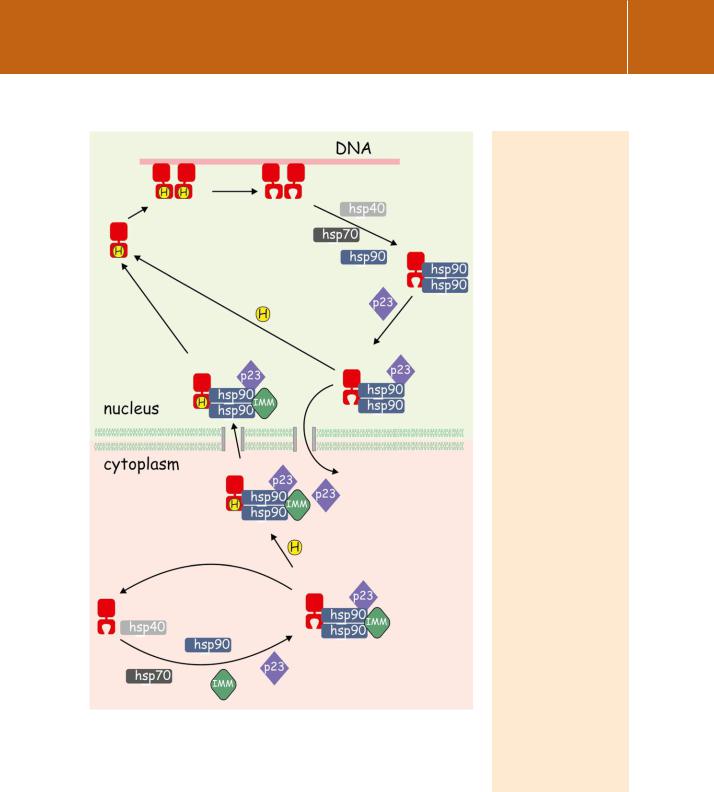
Nuclear receptors
FIG 10.6 A chaperone machine enables cytosolic steroid receptors to bind hormone and enter the nucleus. The sequential recruitment of molecular chaperones, including hsp40, hsp70, and hsp90, with cochaperones, including p23 and an immunophilin (IMM), enables the binding of steroid hormone to the receptor. The complex may then enter the nucleus and release hormone-bound receptor which binds as a dimer to specific DNA sites. (The transcription complex that forms at this stage is not shown.) Finally, receptors may either
be recycled within the nuclear compartment, with the help of the chaperones, or they may be exported or degraded. Based on the glucocorticoid receptor.34, 35
Heat shock proteins.
Here is another example of a chance error that led to an important discovery. Before the recombinant revolution, gene expression was studied by microscopic examination of the expansion (puffing) of bands in
the giant polytene chromosomes of organisms like Drosophila. The expansion is due to the decondensation of chromatin.
Ferruccio Ritossa relates how, as a young graduate student at the University of Pavia in the early 1960s,31 he investigated the type of nucleic acid in the chromosomal puffs of Drosophila salivary glands. A colleague had inadvertently increased the temperature of his incubator and he noticed another puffing pattern. New RNA was synthesized within minutes. The importance of the expressed heat shock proteins as molecular chaperones came later.32
Heat shock proteins also have diverse roles in unstressed cells, affecting folding, packaging, secretion, and degradation of other proteins. Failure of these systems results
in a number of important human diseases.33
283
