
- •SEQUENCE ALIGNMENT
- •Sequence alignment produced with the freely available program ClustalW between two zinc finger
- •WHY DO SIMILARITY SEARCH?
- •WARNING: SIMILARITY NOT
- •BLAST
- •EXAMPLE BLAST QUESTIONS
- •IDENTIFYING SIMILARITY
- •NEEDLEMANWUNSCH
- •NEEDLEMANWUNSCH DETAILS
- •NEEDLEMANWUNSCH
- •ALGORITHM
- •SMITH–WATERMAN ALGORITHM
- •SMITHWATERMAN
- •GLOBAL VS. LOCAL
- •COMPLEXITY
- •OTHER OBSERVATIONS
- •BLAST
- •BLAST is one of the most widely used bioinformatics programs, because it addresses
- •EXAMPLES
- •PROGRAM
- •OUTPUT
- •HOW IT WORKS?
- •BLASTP ALGORITHM (A PROTEIN TO
- •List all of the HSPs in the database whose score is high enough
- •EXTENSIONS
- •EXTENSIONS
- •USES OF BLAST
- •VIDEO LINKS
- •VIDEO LINKS
- •TUTORIALS

LECTURE  #4
#4

SEQUENCE ALIGNMENT
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural,
or evolutionary relationships between the sequences.
Aligned sequences of nucleotide or amino
acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns.
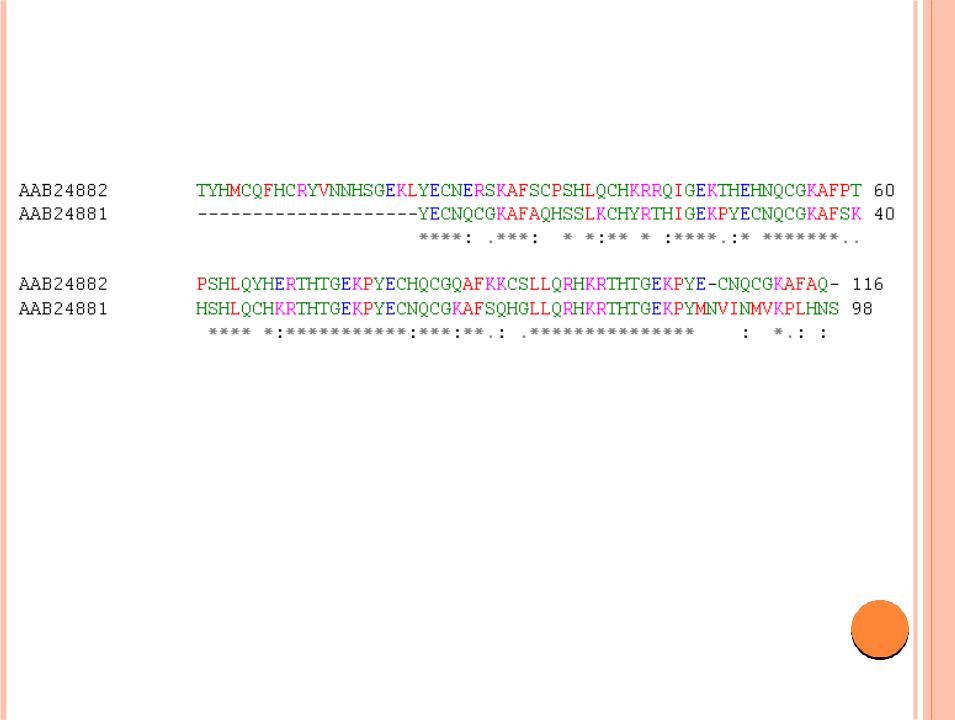
Sequence alignment produced with the freely available program ClustalW between two zinc finger protein sequences publicly available in GenBank

WHY DO SIMILARITY SEARCH?
Similarity indicates conserved function
Human and mouse genes are more than 80% similar at sequence level
But these genes are small fraction of genome
Most sequences in the genome are not recognizably similar
Comparing sequences helps us understand function
Locate similar gene in another species to understand your new gene
Rosetta stone

WARNING: SIMILARITY NOT
TRANSITIVE!
If 1 is “similar” to 2, and 3 is “similar” to 2, is 1 similar to 3?
Not necessarily
AAAAAABBBBBB is similar to AAAAAA and BBBBBB
But AAAAAA is not similar to BBBBBB
“not transitive unless alignments are overlapping”

BLAST
Basic Local Alignment Search Tool
Algorithm for comparing a given sequence against sequences in a database
A match between two sequences is an alignment
Many BLAST databases and web services available

EXAMPLE BLAST QUESTIONS
Which bacterial species have a protein that is related in lineage to a protein whose aminoacid sequence I know?
Where does the DNA I’ve sequenced come from?
What other genes encode proteins that exhibit structures similar to the one I’ve just determined?

IDENTIFYING SIMILARITY
Algorithms to match sequences:
NeedlemanWunsch
Smith Waterman
BLAST

NEEDLEMANWUNSCH
Global alignment algorithm
The algorithm was published in 1970 by Saul B. Needleman and Christian D. Wunsch.
An example: align COELACANTH and PELICAN
Scoring scheme: +1 if letters match, 1 for mismatches, 1 for gaps
COELACANTH COELACANTH P-ELICAN-- -PELICAN--

NEEDLEMANWUNSCH DETAILS
Twodimensional matrix
Diagonal when two letters align
Horizontal when letters paired to gaps
 C
C  O
O
P CP O
 E
E 
L
 I
I
C
 A
A  N
N
 E
E  L
L  A
A  C
C  A
A  N
N  T
T  H
H 
E
E
L
L
A
I
C
C
A
A
N T H
N - -

NEEDLEMANWUNSCH
In reality, each cell of matrix contains score and pointer
Score is derived from scoring scheme (1 or +1 in our example)
Pointer is an arrow that points up, left, or diagonal
After initializing matrix, compute the score and arrow for each cell
