
Robert I. Kabacoff - R in action
.pdf326 |
CHAPTER 13 Generalized linear models |
|
|||
Age |
0.022740 |
0.004024 |
5.65 |
1.6e-08 |
*** |
Trtprogabide -0.152701 |
0.047805 |
-3.19 |
0.0014 |
** |
|
--- |
|
|
|
|
|
Signif. codes: |
0 '***' 0.001 '**' 0.01 '*' |
0.05 '.' 0.1 ' ' 1 |
|||
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 2122.73 on 58 degrees of freedom
Residual deviance: 559.44 on 55 degrees of freedom
AIC: 850.7
Number of Fisher Scoring iterations: 5
The output provides the deviances, regression parameters, standard errors, and tests that these parameters are 0. Note that each of the predictor variables is significant at the p<0.05 level.
13.3.1Interpreting the model parameters
The model coefficients are obtained using the coef() function, or by examining the Coefficients table in the summary() function output:
> coef(fit) |
|
|
|
(Intercept) |
Base |
Age |
Trtprogabide |
1.9488 |
0.0227 |
0.0227 |
-0.1527 |
In a Poisson regression, the dependent variable being modeled is the log of the conditional mean loge(λ) . The regression parameter 0.0227 for Age indicates that a oneyear increase in age is associated with a 0.03 increase in the log mean number of seizures, holding baseline seizures and treatment condition constant. The intercept is the log mean number of seizures when each of the predictors equals 0. Because you can’t have a zero age and none of the participants had a zero number of baseline seizures, the intercept isn’t meaningful in this case.
It’s usually much easier to interpret the regression coefficients in the original scale of the dependent variable (number of seizures, rather than log number of seizures). To accomplish this, exponentiate the coefficients:
> exp(coef(fit)) |
|
|
|
(Intercept) |
Base |
Age |
Trtprogabide |
7.020 |
1.023 |
1.023 |
0.858 |
Now you see that a one-year increase in age multiplies the expected number of seizures by 1.023, holding the other variables constant. This means that increased age is associated with higher numbers of seizures. More importantly, a one-unit change in Trt (that is, moving from placebo to progabide) multiplies the expected number of seizures by 0.86. You’d expect a 20 percent decrease in the number of seizures for the drug group compared with the placebo group, holding baseline number of seizures and age constant.
It’s important to remember that, like the exponeniated parameters in logistic
328 |
CHAPTER 13 Generalized linear models |
family="poisson" with family="quasipoisson". Doing so is analogous to our approach to logistic regression when overdispersion is present.
> fit.od <- glm(sumY ~ Base + Age + Trt, data=breslow.dat, family=quasipoisson())
> summary(fit.od)
Call:
glm(formula = sumY ~ Base + Age + Trt, family = quasipoisson(), data = breslow.dat)
Deviance Residuals: |
|
|
|
|
||
Min |
1Q |
Median |
3Q |
Max |
|
|
-6.057 -2.043 -0.940 |
0.793 |
11.006 |
|
|
||
Coefficients: |
|
|
|
|
|
|
|
Estimate Std. Error t value Pr(>|t|) |
|
||||
(Intercept) |
|
1.94883 |
0.46509 |
4.19 |
0.00010 |
*** |
Base |
|
0.02265 |
0.00175 |
12.97 |
< 2e-16 *** |
|
Age |
|
0.02274 |
0.01380 |
1.65 |
0.10509 |
|
Trtprogabide -0.15270 |
0.16394 |
-0.93 |
0.35570 |
|
||
--- |
|
|
|
|
|
|
Signif. codes: |
0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 |
|||||
(Dispersion parameter for quasipoisson family taken to be 11.8)
Null deviance: 2122.73 on 58 degrees of freedom
Residual deviance: 559.44 on 55 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5
Notice that the parameter estimates in the quasi-Poisson approach are identical to those produced by the Poisson approach. The standard errors are much larger, though. In this case, the larger standard errors have led to p-values for Trt (and Age) that are greater than 0.05. When you take overdispersion into account, there’s insufficient evidence to declare that the drug regimen reduces seizure counts more than receiving a placebo, after controlling for baseline seizure rate and age.
Please remember that this example is used for demonstration purposes only. The results shouldn’t be taken to imply anything about the efficacy of progabide in the real world. I’m not a doctor—at least not a medical doctor—and I don’t even play one on TV.
We’ll finish this exploration of Poisson regression with a discussion of some important variants and extensions.
13.3.3Extensions
R provides several useful extensions to the basic Poisson regression model, including models that allow varying time periods, models that correct for too many zeros, and robust models that are useful when data includes outliers and influential observations. I’ll describe each separately.
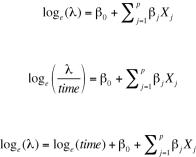
Poisson regression |
329 |
POISSON REGRESSION WITH VARYING TIME PERIODS
Our discussion of Poisson regression has been limited to response variables that measure a count over a fixed length of time (for example, number of seizures in an eight-week period, number of traffic accidents in the past year, number of pro-social behaviors in a day). The length of time is constant across observations. But you can fit Poisson regression models that allow the time period to vary for each observation. In this case, the outcome variable is a rate.
To analyze rates, you must include a variable (for example, time) that records the length of time over which the count occurs for each observation. You then change your model from
to
or equivalently to
To fit this new model, you use the offset option in the glm() function. For example, assume that the length of time that patients participated post-randomization in the Breslow study varied from 14 days to 60 days. You could use the rate of seizures as the dependent variable (assuming you had recorded time for each patient in days), and fit the model
fit <- glm(sumY ~ Base + Age + Trt, data=breslow.dat, offset= log(time), family=poisson())
where sumY is the number of seizures that occurred post-randomization for a patient during the time the patient was studied. In this case you’re assuming that rate doesn’t vary over time (for example, 2 seizures in 4 days is equivalent to 10 seizures in 20 days).
ZERO-INFLATED POISSON REGRESSION
There are times when the number of zero counts in a dataset is larger than would be predicted by the Poisson model. This can occur when there’s a subgroup of the population that would never engage in the behavior being counted. For example, in the Affairs dataset described in the section on logistic regression, the original outcome variable (affairs) counted the number of extramarital sexual intercourse experience participants had in the past year. It’s likely that there’s a subgroup of faithful marital partners who would never have an affair, no matter how long the period of time studied. These are called structural zeros (primarily by the swingers in the group).
In such cases, you can analyze the data using an approach called zero-inflated Poisson regression. The approach fits two models simultaneously—one that predicts who would or would not have an affair, and the second that predicts how many affairs a participant

330 |
CHAPTER 13 Generalized linear models |
would have if you excluded the permanently faithful. Think of this as a model that combines a logistic regression (for predicting structural zeros) and a Poisson regression model (that predicts counts for observations that aren’t structural zeros). Zero-inflated Poisson regression can be fit using the zeroinfl() function in the pscl package.
ROBUST POISSON REGRESSION
Finally, the glmRob() function in the robust package can be used to fit a robust generalized linear model, including robust Poisson regression. As mentioned previously, this can be helpful in the presence of outliers and influential observations.
Going further
Generalized linear models are a complex and mathematically sophisticated subject, but many fine resources are available for learning about them. A good, shor t introduction to the topic is Dunteman and Ho (2006). The classic (and advanced) text on generalized linear models is provided by McCullagh and Nelder (1989). Comprehensive and accessible presentations are provided by Dobson and Barnett (2008) and Fox (2008). Faraway (2006) and Fox (2002) provide excellent introductions within the context of R.
13.4 Summary
In this chapter, we used generalized linear models to expand the range of approaches available for helping you to understand your data. In particular, the framework allows you to analyze response variables that are decidedly non-normal, including categorical outcomes and discrete counts. After briefly describing the general approach, we focused on logistic regression (for analyzing a dichotomous outcome) and Poisson regression (for analyzing outcomes measured as counts or rates).
We also discussed the important topic of overdispersion, including how to detect it and how to adjust for it. Finally, we looked at some of the extensions and variations that are available in R.
Each of the statistical approaches covered so far has dealt with directly observed and recorded variables. In the next chapter, we’ll look at statistical models that deal with latent variables—unobserved, theoretical variables that you believe underlie and account for the behavior of the variables you do observe. In particular, you’ll see how you can use factor analytic methods to detect and test hypotheses about these unobserved variables.


332 |
CHAPTER 14 Principal components and factor analysis |
In contrast, exploratory factor analysis (EFA) is a collection of methods designed to uncover the latent structure in a given set of variables. It looks for a smaller set of underlying or latent constructs that can explain the relationships among the observed or manifest variables. For example, the dataset Harman74.cor contains the correlations among 24 psychological tests given to 145 seventhand eighth-grade children. If you apply EFA to this data, the results suggest that the 276 test intercorrelations can be explained by the children’s abilities on four underlying factors (verbal ability, processing speed, deduction, and memory).
The differences between the PCA and EFA models can be seen in figure 14.1. Principal components (PC1 and PC2) are linear combinations of the observed variables (X1 to X5). The weights used to form the linear composites are chosen to maximize the variance each principal component accounts for, while keeping the components uncorrelated.
In contrast, factors (F1 and F2) are assumed to underlie or “cause” the observed variables, rather than being linear combinations of them. The errors (e1 to e5) represent the variance in the observed variables unexplained by the factors. The circles indicate that the factors and errors aren’t directly observable but are inferred from the correlations among the variables. In this example, the curved arrow between the factors indicates that they’re correlated. Correlated factors are common, but not required, in the EFA model.
The methods described in this chapter require large samples to derive stable solutions. What constitutes an adequate sample size is somewhat complicated. Until recently, analysts used rules of thumb like “factor analysis requires 5–10 times as many subjects as variables.” Recent studies suggest that the required sample size depends on the number of factors, the number of variables associated with each factor, and how well the set of factors explains the variance in the variables (Bandalos and Boehm-Kaufman, 2009). I’ll go out on a limb and say that if you have several hundred observations, you’re probably safe. In this chapter, we’ll look at artificially small problems in order to keep the output (and page count) manageable.
Figure 14.1 Comparing principal components and factor analysis models. The diagrams show the observed variables (X1 to X5), the principal components (PC1, PC2), factors (F1, F2), and errors (e1
to e5).

Principal components and factor analysis in R |
333 |
We’ll start by reviewing the functions in R that can be used to perform PCA or EFA and give a brief overview of the steps involved. Then we’ll work carefully through two PCA examples, followed by an extended EFA example. A brief overview of other packages in R that can be used for fitting latent variable models is provided at the end of the chapter. This discussion includes packages for confirmatory factor analysis, structural equation modeling, correspondence analysis, and latent class analysis.
14.1 Principal components and factor analysis in R
In the base installation of R, the functions for PCA and EFA are princomp() and factanal(), respectively. In this chapter, in this chapter, we’ll focus on functions provided in the psych package. They offer many more useful options than their base counterparts. Additionally, the results are reported in a metric that will be more familiar to social scientists and more likely to match the output provided by corresponding programs in other statistical packages such as SAS and SPSS.
The psych package functions that are most relevant here are listed in table 14.1. Be sure to install the package before trying the examples in this chapter.
Table 14.1 Useful factor analytic functions in the psych package
Function |
Description |
|
|
principal() |
Principal components analysis with optional rotation |
fa() |
Factor analysis by principal axis, minimum residual, weighted least squares, or |
|
maximum likelihood |
fa.parallel() |
Scree plots with parallel analyses |
factor.plot() |
Plot the results of a factor or principal components analysis |
fa.diagram() |
Graph factor or principal components loading matrices |
scree() |
Scree plot for factor and principal components analysis |
|
|
EFA (and to a lesser degree PCA) are often confusing to new users. The reason is that they describe a wide range of approaches, and each approach requires several steps (and decisions) to achieve a final result. The most common steps are as follows:
1Prepare the data. Both PCA and EFA derive their solutions from the correlations among the observed variables. Users can input either the raw data matrix or the correlation matrix to the principal() and fa() functions. If raw data is input, the correlation matrix will automatically be calculated. Be sure to screen the data for missing values before proceeding.
2Select a factor model. Decide whether PCA (data reduction) or EFA (uncovering latent structure) is a better fit for your research goals. If you select an EFA approach, you’ll also need to choose a specific factoring method (for example, maximum likelihood).

334 |
CHAPTER 14 Principal components and factor analysis |
3Decide how many components/factors to extract.
4Extract the components/factors.
5Rotate the components/factors.
6Interpret the results.
7Compute component or factor scores.
In the remainder of this chapter, we’ll carefully consider each of the steps, starting with PCA. At the end of the chapter, you’ll find a detailed flow chart of the possible steps in PCA/EFA (figure 14.7). The chart will make more sense once you’ve read through the intervening material.
14.2 Principal components
The goal of PCA is to replace a large number of correlated variables with a smaller number of uncorrelated variables while capturing as much information in the original variables as possible. These derived variables, called principal components, are linear combinations of the observed variables. Specifically, the first principal component
PC1 = a1X1 + a2X2 + … + akX k
is the weighted combination of the k observed variables that accounts for the most variance in the original set of variables. The second principal component is the linear combination that accounts for the most variance in the original variables, under the constraint that it’s orthogonal (uncorrelated) to the first principal component. Each subsequent component maximizes the variance accounted for, while at the same time remaining uncorrelated with all previous components. Theoretically, you can extract as many principal components as there are variables. But from a practical viewpoint, you hope that you can approximate the full set of variables with a much smaller set of components. Let’s look at a simple example.
The dataset USJudgeRatings contains lawyers’ ratings of state judges in the US Superior Court. The data frame contains 43 observations on 12 numeric variables. The variables are listed in table 14.2.
Table 14.2 Variables in the USJudgeRatings dataset
Variable |
Description |
Variable |
Description |
|
|
|
|
CONT |
Number of contacts of lawyer with judge |
PREP |
Preparation for trial |
INTG |
Judicial integrity |
FAMI |
Familiarity with law |
DMNR |
Demeanor |
ORAL |
Sound oral rulings |
DILG |
Diligence |
WRIT |
Sound written rulings |
CFMG |
Case flow managing |
PHYS |
Physical ability |
DECI |
Prompt decisions |
RTEN |
Wor thy of retention |
|
|
|
|
Principal components |
335 |
From a practical point of view, can you summarize the 11 evaluative ratings (INTG to RTEN) with a smaller number of composite variables? If so, how many will you need and how will they be defined? Because our goal is to simplify the data, we’ll approach this problem using PCA. The data are in raw score format and there are no missing values. Therefore, your next decision is deciding how many principal components you’ll need.
14.2.1Selecting the number of components to extract
Several criteria are available for deciding how many components to retain in a PCA. They include:
■Basing the number of components on prior experience and theory
■Selecting the number of components needed to account for some threshold cumulative amount of variance in the variables (for example, 80 percent)
■Selecting the number of components to retain by examining the eigenvalues of the k x k correlation matrix among the variables
The most common approach is based on the eigenvalues. Each component is associated with an eigenvalue of the correlation matrix. The first PC is associated with the largest eigenvalue, the second PC with the second-largest eigenvalue, and so on. The Kaiser–Harris criterion suggests retaining components with eigenvalues greater than 1. Components with eigenvalues less than 1 explain less variance than contained in a single variable. In the Cattell Scree test, the eigenvalues are plotted against their component numbers. Such plots will typically demonstrate a bend or elbow, and the components above this sharp break are retained. Finally, you can run simulations, extracting eigenvalues from random data matrices of the same size as the original matrix. If an eigenvalue based on real data is larger than the average corresponding eigenvalues from a set of random data matrices, that component is retained. The approach is called parallel analysis (see Hayton, Allen, and Scarpello, 2004 for more details).
You can assess all three eigenvalue criteria at the same time via the fa.parallel() function. For the 11 ratings (dropping the CONT variable), the necessary code is as follows:
library(psych)
fa.parallel(USJudgeRatings[,-1], fa="PC", ntrials=100, show.legend=FALSE, main="Scree plot with parallel analysis")
This code produces the graph shown in figure 14.2. The plot displays the scree test based on the observed eigenvalues (as straight-line segments and x’s), the mean eigenvalues derived from 100 random data matrices (as dashed lines), and the eigenvalues greater than 1 criteria (as a horizontal line at y=1).
All three criteria suggest that a single component is appropriate for summarizing this dataset. Your next step is to extract the principal component using the principal() function.
