
Solid-Phase Synthesis and Combinatorial Technologies
.pdf
|
|
|
|
|
|
4.2 |
COMBINATORIAL LIBRARIES 153 |
|||
|
N |
O |
|
|
|
CN |
|
O |
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
N |
|
|
|
||
R1 |
|
|
R2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
N |
|
N |
||
|
L13 |
|
|
|
|
|
|
|||
|
R1 |
|
L24 |
|
R |
|
|
|
||
|
|
|
|
|
N |
|
O |
|||
|
|
|
|
|
|
2 |
|
|||
|
|
|
R3 |
|
|
|
R1 |
|
|
R2 |
|
N |
O |
|
|
|
|
L15 |
|||
|
|
|
|
|
|
R3 |
||||
|
|
|
|
|
O |
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
R1 |
N |
|
|
|
|
|
|
|
|
|
|
|
R1 |
|
L12 |
|
R2 |
|
|
N N |
|
|
H |
|
|
|
|
|
|
|||
|
L21 |
R |
|
|
|
|
R1 |
|
|
R |
|
|
2 |
|
|
|
|
|
|
|
2 |
|
|
|
|
|
|
|
|
|
|
L16 |
|
R3 |
(n) |
|
|
|
|
|
O |
O |
R3 |
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
N |
|
N |
O |
|
|
|
|
|
|
|
H |
|
|
|
|
|
|
|
|
|
||
|
|
|
N |
|
R3 |
R1 |
L17 |
|
R2 |
|
R1 |
|
|
|
|
|
|||||
L20 |
R2 |
|
|
|
R3 |
R4 |
|
|||
|
N |
N |
|
|
|
|||||
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
O |
N |
|
|
|
|
R1 |
L19 |
R2 |
|
|
O |
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
R1 |
|
L18 |
|
|
R2 |
L12
L13
L14
L15
L16
L17
L18
L19
L20
L21
R1: 32 acetophenones, R2: 40 aromatic aldehydes library size: 32x40=1,280 discrete chalcones
L12 reacted with NH2OH library size: 1,280x1=1,280 discrete isoxazolines
L12 reacted with 3-aminocrotononitrile |
library size: 1,280x1=1,280 discrete pyridines |
||
L12 reacted with 6-NH2-1,3-diMeuracyl |
library size: 1,280x1=1,280 discrete byciclic compounds |
||
L12 reacted with 6 phenylhydrazines |
library size: 1,280x6=7,680 discrete pyrazolines |
||
L12 reacted with 6 acetoacetanilides |
library size: 1,280x6=7,680 discrete cyclohexenones |
||
L12 (320 cpds) with 40 acetoacetamides |
library size: 320x40=12,800 discrete cyclohexenones |
||
L12 with 6 aminobenzimidazoles |
library size: 1,280x6=7,680 discrete tricyclic compounds |
||
L12 with 6 cyclic ketones and NH3 |
library size: 1,280x6=7,680 discrete byciclic compounds |
||
L12 (80 cpds) reacted with 16 isatins and 20 amino acids library size: 80x16x20=1,280 discrete spiro compounds
Figure 4.13 Structure of a solution-phase primary modular library L12 based on a chalcone core and of several derived libraries L13–L21.
The combinatorial modification of yohimbinic acid, a natural alkaloid, was reported by Atuegbu et al. (48), and the structure of the derived SP pool library L22 is shown in Fig. 4.14. The library was made by 36 × 22 = 792 compounds as 22 pools of 36 compounds, and the monomer sets used were L-α-amino acids (R1) and carboxylic

154 COMBINATORIAL TECHNOLOGIES: BASIC PRINCIPLES
|
|
R1: from 36 amino acids |
|
N |
N |
R2: from 22 carboxylic acids |
|
H |
|||
H H |
|
||
O |
H |
library size: 36x22=792 difunctionalized |
|
H |
yohimbamides |
||
N |
|
||
|
|
||
H2N |
|
prepared as 22 pools of 36 compounds |
|
R1 |
O O R2 |
||
|
L22 O
Figure 4.14 Structure of an SP yohimbinic acid decoration library L22.
acids (R2). The biological evaluation of the library was not reported. The commercially available natural scaffold is known to possess a wide range of biological activities, and the possibility to further expand the two monomer sets make such a primary library a likely source for new biologically active analogues. There is a real possibility of building a preliminary structure activity relationship (SAR) and of finding substituents that favor different biological activities providing that the decoration of the natural product diversifies the library components enough.
Another decoration pool library L23 was reported by Nestler (49), who presented the appendage of a peptidic chain to the two hydroxylic functions of a steroid scaffold (Fig. 4.15). The library was made up of 10 × 10 × 10 × 10 = 10,000 individuals using a chemical encoding method (39) and L-α-amino acids as monomer sets (R1–R4). The assay of the library as a source of artificial two-armed receptors for enkephalin-related peptides produced positives with micromolar affinity. Much larger libraries could be obtained by simply increasing the monomer sets and the length of the two arms; this could lead to a primary library of peptide-binding artificial receptors; similar scaffolds have been repeatedly exploited for combinatorial purposes (50–52).
|
|
Me |
|
|
Me |
|
Me |
|
|
|
HH |
|
O |
O |
|
H |
Gly |
|
Gly |
|
X1 |
L23 |
X3 |
X2 |
X4 |
|
O |
|
O |
H
N
O
X1-X4: 10 L-α-amino acids: Ala, Val, Leu, Phe, Pro, Ser, Thr, Lys, Glu, Asp
library size: 10x10x10x10=10,000 decorated cholates prepared as a bead-based encoded library
Figure 4.15 Structure of an SP steroid decoration library L23.

4.2 COMBINATORIAL LIBRARIES 155
Quinic acid was decorated on the C1-carboxylic acid and on the C3, C4, C5-hydroxyls by Phoon and Abell (53) as in Fig. 4.16, producing two small arrays L24 (ten monoester-monoamide compounds) and L25 (six triester-monoamide compounds). The reliability and the robustness of both the scaffold and of the acylation/esterifications ensure an expansion of quinate-based libraries, which has already been reported for shikimate-based libraries (54).
The natural scaffold, or a scaffold resembling a natural product, can also be built during the synthesis of the library. An example presented by Nielsen and Lyngsoe (55) involved the synthesis of a library L26 inspired by the structure of balanol (Fig. 4.17). A disconnection study revealed α-amino alcohols Al, aromatic dicarboxylic acids Ar, and aromatic carboxylic acids R1 as monomer sets to mimic the natural constituents of balanol, and the small library L26 was prepared. It was made up of 2 × 4 × 4 = 32 compounds prepared as four pools of eight compounds that could be expanded by using enlarged monomer sets to produce primary balanol-biased combinatorial libraries. Introduction of modified building blocks (aliphatic diacids or monoacids, acyclic or aromatic amino alcohols, etc.) could lead, after substantial assessment of the chemistry, to a modular library of different balanol-inspired compounds with a high level of diversity.
A more complex combinatorial strategy based on a natural scaffold was reported by Nicolaou et al. (56) with the synthesis of an epothilone-based SP library L27 (Fig. 4.18). This biased library was prepared as a 3 × 3 × 5 = 45-member collection, but the final reaction vessels could, theoretically contain 4 different isomeric desoxyepothilones (Fig. 4.18), thus leading to a total number of 45 × 4 = 180 components. The library was prepared using a radiofrequency encoding technique (42), and the final
|
O |
|
|
O |
|
R1 |
W O |
R1 |
|
W O |
||||
|
N |
|
|
N |
|
|
|
H |
|
|
H |
|
|
|
|
O |
O |
O |
|
|
|
|||
|
|
|
|
|
HO |
O |
R |
R3 O |
O R2 |
|
|
|
2 |
O R3 |
|
OH |
|
|
|
|
L24 |
|
L25 |
O |
|
|
6 discretes |
||
|
10 discretes |
|
|
|
|
|
|
|
|
R1: 3 primary amines (n-propyl, benzyl and cyclohexyl)
R2: 5 carboxylic acids (acetic, benzoic, 4-pyridyl, phenylacetic and acrylic)
R3: 2 carboxylic acids (acetic and benzoic)
Figure 4.16 Structure of two quinicacid-inspired SP small arrays L24 and L25.

156 COMBINATORIAL TECHNOLOGIES: BASIC PRINCIPLES
|
|
HOOC |
|
|
OH |
|
|
|
|
|
OH O |
|
|
|
|
||
|
|
|
|
|
|
|
Al |
|
|
|
|
|
|
N |
NH2 |
HOOC |
NH2 |
|
|
|
|
|
Al=aliphatic |
|||
|
|
O O |
|
H |
|
|||
|
O |
|
|
|
|
|||
|
|
|
|
|
|
|||
|
O |
H |
H |
|
|
OH |
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
N |
NH |
|
|
|
|
|
|
R1 |
|
|
|
|
|
|
|
||
H |
|
|
|
HOOC |
|
HOOC |
|
|
|
O |
|
|
|
|
|||
|
|
|
|
|
|
|
||
|
OH |
|
|
HOOC |
|
|
||
|
|
|
OH |
|
|
|||
|
Balanol |
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
HOOC |
Ar |
|
|
|
|
|
|
|
COOH |
|
|
|
|
|
HOOC |
O O |
|
|
|
|
|
|
|
|
|
H |
|
|
|
|
|
|
|
|
H |
|
|
|
|
H |
|
R1 |
Ar: 2 aromatic diacids |
|
|
|
|
Ar |
|
Al: 4 aliphatic amino alcohols |
|
||||
|
O N |
|
|
|
||||
HOOC |
Al |
|
|
R1: 4 benzoic acids |
|
|
||
|
O |
O |
|
|
|
|
||
|
|
|
|
|
|
|
||
|
|
L26 |
|
|
library size: 2x4x4=32 individuals |
|
||
prepared as four pools of 8 compounds
Figure 4.17 Structure of an SP balanol-inspired library L26.
cleavage solutions from each vessel were purified to give pure single isomers that were assayed to obtain a detailed SAR (56). The monomer sets consisted of three protected hydroxy aldehydes (R1), three protected hydroxy keto acids (R2), and five heterocyclic secondary alcohols (R3) prepared using published multistep routes, and the expansion of these sets would require significant synthetic effort. Once such an ambitious synthetic scheme has been set up, it would be desirable to move away from the initially used biased monomer sets and to introduce more diverse commercially available monomers to build larger, less epothilone-like libraries with the same synthetic strategy that could be used as primary libraries to search for other biological activities. This example shows that even complex natural scaffolds can be dissected and reconstructed using appropriate reactions and monomer sets. The balance between the appeal of the designed library and the resources/effort needed to set up the library synthesis should drive the strategic decision to go for the synthesis or not.
4.2.4 Biological Libraries
Libraries produced by microorganisms or through biochemical techniques have been frequently used to find either peptidic or oligonucleotidic sequences that bind certain receptors, or enzymes, and/or possess catalytic properties (catalytic antibodies and ribozymes, among others). Biological libraries can also be produced by combinatorial
|
|
|
|
|
4.2 |
COMBINATORIAL LIBRARIES |
157 |
|
R1 |
|
|
R1: from |
OH |
R1 |
O , as OH |
O |
|
OH |
|
|
R3 |
|
|
|
|
|
|
|
O |
R2: from |
|
|
COOH |
|
|
|
|
|
|
|
, as |
COOH |
||
|
R2 O |
|
|
|
R2 |
|
||
O |
|
|
O |
|
O |
|
||
L27
Library size: 3x3x5=45 samples R3: from (four stereoisomers each, 180 individuals)
prepared as 45 radioencoded discretes
prep. TLC
R1
OH |
R3 |
O +
O R2 O
L27a
R1
OH
O |
|
|
R3 + |
|
|||
|
|
|

 O
O
R2 O
S
OH
, as
 R3
R3
OH
|
R1 |
OH |
R3 |
|
O |
O R2 O
L27b
R1
OH
O |
|
R3 |
|
 O
O
R2
O
L27c |
L27d |
Figure 4.18 Structure of an SP epothilone-inspired library L27.
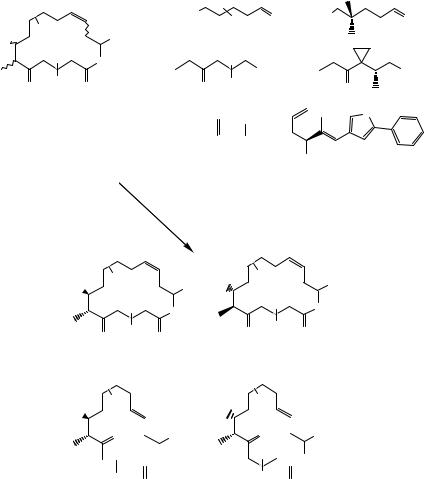
modifications of chemical scaffolds performed by isolated enzymes, whole microorganisms, and even by the so-called combinatorial biosynthesis, which alters the enzymatic machinery responsible for the synthesis of natural products, thus allowing the preparation of combinatorial biosynthetic analogues. Some biological libraries are reported in Figs. 4.19–4.22.
An application of phage display libraries to the identification of peptidic sequences that selectively target tumor blood vessels was reported by Arap et al. (57). Three cyclic peptide libraries were produced by inserting the corresponding degenerate oligonucleotide sequences into a vector (fUSE 5), then transforming MC1061 cells by

158 COMBINATORIAL TECHNOLOGIES: BASIC PRINCIPLES
X |
X |
X |
X |
X |
X |
Cys |
X |
Cys |
X |
Cys X |
Cys |
|
|
|
L28 |
|
X=20 natural L-α-amino acids |
|||
|
|
phage library size: |
||||||
|
|
|
|
|
|
|||
20x20x20x20x20x20x20x20x20=512,000,000,000 cyclic peptides |
|
|
|
|||||
X |
X |
X |
X |
X |
X |
X |
X |
X |
Cys |
X |
X |
Cys |
Cys |
X |
Cys |
||
|
|
L29 |
|
|
L30 |
|
||
phage library size: |
|
phage library size: |
||||||
20x20x20x20x20x20x20=1,280,000,000 cyclic peptides |
20x20x20x20x20=3,200,000 cyclic peptides |
|||||||
Figure 4.19 Structure of phage display S–S cyclic peptide libraries L28–L30.
electroporation. The libraries produced contained two terminal cysteines to provide S–S cyclization of the peptides: the CX3CX3CX3C library (L28), the CX7C library (L29), and the CX5C library (L30) (Fig. 4.19). Three distinct structural motifs were recovered after in vivo selection in tumor-bearing mice; NGR (in CNGRCVSGCAGRC from L28), RGD (in CDCRGDCFC from L29), and GSL (in CGSLVRC from L30). All three of them delivered peptidic sequences preferentially to the tumor and seemed to bind to different tumor receptors. Their use as drug delivery systems for the development of targeted chemotherapy looks extremely promising. The usefulness of phage display libraries in producing enormous numbers of peptidic sequences of various lengths combined with the possibility of tailoring the structures of the displayed population and the commercial availability of several phage systems makes this biological library source extremely appealing. Production of biochemical reagents (e.g., antibodies, diagnostics, and vaccines) or tools (e.g., to validate targets via their inhibition and to produce peptidic ligands) and gathering structural information about receptors by finding peptide agonists or antagonists have all been reported. Undoubtedly, the exploitation of phage libraries will increase and significant breakthroughs are to be expected in the future.
An example of deoxyribozyme (deoxyoligonucleotidic enzymatic activity) selection applied to the isolation of a broad specificity RNA-cleaving activity was reported by Santoro and Joyce (58). A specific construct was built up by assembling a 5′-biotin moiety (A), attached by a short ODN spacer (B), to a target 12-mer ribonucleotide containing the target cleavage sequence (C) and a 50-mer random ODN (E). This latter sequence was surrounded by fixed-sequence ODN spacers (D) on the 3′ and 5′ ends (Fig. 4.20). The corresponding library L31 (1014 ON–ODN components) was applied to a streptavidin-coated support and bound tightly to it via the biotin–streptavidin recognition. The few RNA-cleaving DNA sequences released only the cleaved constructs, which, after 10 rounds of polymerase chain reaction (PCR) amplification, eventually produced two catalytic motifs (DNA 1 and 2, Fig. 4.20) which were thoroughly studied and refined. Many other reports of ribozymes and deoxyribozymes have been reported, as the use of PCR amplification to select ON–ODN aptamers that bind to receptors/small molecules.

|
|
|
|
|
|
|
|
|
|
|
4.2 COMBINATORIAL LIBRARIES 159 |
||||
|
|
D |
C (ON 12-mer) |
|
|
|
B |
A |
|
|
|
|
|
|
|
|
|
|
UAGAGAUCAAUG |
library size: 1014 ON(C)-ODN(B,D,E) individuals |
|||||||||||
|
|
|
|
||||||||||||
|
|
|
|
|
|
|
|
|
prepared as mixture, then amplified ten times by |
||||||
|
|
|
|
|
|
|
|
3' |
PCR only on selected sequences by reverse transcriptase |
||||||
E (ODN randomized 50-mer) |
|
D |
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
L31 |
|
|
|
|
|
|
|
C (ON 11-mer) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
D |
|
|
|
||
|
|
|
|
|
|
|
|
|
|
UAGAGAUCAAU |
|
|
|
||
|
|
|
selection |
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3' |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C (ON 12-mer) |
|
|
|
|
|
E (RNA-cleaving DNA |
1) |
|
|
D |
||
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
D |
|
B |
A |
|
|
in solution |
|
|
|
|
|||
|
|
|
UAGAGAUCAAUG |
|
|
|
|
+ |
D |
C (ON 5-mer) |
|
|
|
|
|
|
|
|
|
|
|
|
|
3' |
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
E (ODN randomized 50-mer) |
|
D |
|
UAGAG |
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|||||||
non RNA-cleaving DNA sequences |
|
|
|
|
|
3' |
|||||||||
|
|
|
support-bound |
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
E (RNA-cleaving DNA |
2) |
|
|
D |
||
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
||||
in solution
PCR amplification
50-member ODNs
DNA 1, DNA 2
Figure 4.20 Structure of an SP ON/ODN library L31.
The generation of biased libraries from synthetic or natural precursors using either purified enzymes or whole microorganisms was reported by Khmelnitsky et al. (59) adopting the so-called combinatorial biocatalysis approach. The library L32, based on (±)-(2-endo,3-exo)-bicyclo[2.2.2]oct-5-ene-2,3-dimethanol (BOD), was obtained by submitting BOD to a panel of multistep enzymatic transformations. A total of 1222 compounds were obtained and some of the representative structures are shown in Fig. 4.21. This approach allows the generation of very diverse and complex compounds from a common scaffold employing mild enzymatic transformations that can also accommodate sensitive substrates. These transformations are often stereoselective and regioselective, so that complex derivatives may be obtained without protection/deprotection steps. A lot of effort is required to process the reaction samples and to isolate, purify, and structurally characterize the final library components. However, this strategy is particularly useful when a lead compound with a complex structure, which has already proven its value for a specific application, must be optimized for progression as a drug candidate.
Combinatorial biosynthesis is an approach aimed at the modification of the cellular machinery involved in the biosynthetic pathways that produce natural products. There are several groups active in the field, and an example reported by McDaniel et al. (60)

160
HO
HO
COMBINATORIAL TECHNOLOGIES: BASIC PRINCIPLES
OH
BOD  OH
OH
L32 library size: 1222 individuals
prepared as discretes submitting BOD to multistep enzymatic biotransformations
including halohydration, glycosylation and acylation
|
|
selected library compounds: |
|
|
|
|
|
OH |
OH |
|
O |
HO |
O |
|
|
|
|
||
|
O |
|
O |
OH |
|
Cl |
OH |
|
|
|
|
|
||
|
|
|
OH |
|
|
OH |
|
|
|
Cl |
|
|
|
|
|
|
|
|
|
|
|
|
|
N |
|
|
|
O |
|
|
|
O |
O |
O |
|
|
|
|
|
O |
O |
O |
|
|
|
|
|
||
|
|
O |
|
|
|
O |
|
|
|
|
OH |
|
|
|
|
|
O |
|
|
|
|
Cl |
|
Figure 4.21 Structure of a combinatorial biocatalysis BOD-derived library L32.
demonstrates the generation of rationally engineered biosynthesis of polyketides. A scheme highlighting the current possibilities offered by biosynthetic manipulation including the biosynthetic modifications involved and the intermediates and the final compound produced is shown in Fig. 4.22. This field has enormous potential and will benefit from efforts aimed at making biosynthetic pathways other than the polyketide available to the armamentarium of combinatorial chemistry.
4.2.5 Materials Science Libraries
Libraries composed of inorganic components, normally referred to as materials science libraries, surely represent one of the most intriguing and interesting class of combinatorial libraries. Since their recent appearance in combinatorial chemistry, they have been applied to many problems in the field of materials science, and an example is

4.2 COMBINATORIAL LIBRARIES 161
Engineered polyketide biosynthesis
Manipulation of the following steps currently feasible:
CHAIN LENGTH of the product
KETOREDUCTION (yes or no)
1st RING STEREOSPECIFICITY
1st RING AROMATIZATION
2nd RING CYCLIZATION (yes or no)
|
|
|
|
combinations of the above |
|
|
|
OH O |
O |
|
OH |
OH O |
O O |
|
O |
O |
O |
or |
|
O |
or |
O |
O |
|
|
||||||
HO |
|
HO |
|
|
HO |
|
|
|
OH |
O O |
|
OH |
O O |
|
|
|
|
O |
O |
or |
O |
|
|
|
|
|
|
HO |
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
uncatalyzed end reaction |
|
|
|
RATIONALLY ENGINEERED POLYKETIDES
Figure 4.22 Combinatorial biosynthesis: manipulation of the aromatic polyketide pathway.
reported in Fig. 4.23, which will be used to briefly define the properties and opportunities provided by these libraries.
A library L33 made up of different compositions and stoichiometries of LnxMy- CoOz (Ln = La or Y, M = Ba, Sr, Ca, or Pb) was reported by Briceno et al. (61) as a source of new magnetoresistant materials. It was composed of 128 discretes (Fig. 4.23) and was prepared in duplicate using a combination of thin-film deposition and physical masking to vary the individual components. Each library copy was analyzed for magnetoresistance using a different procedure. A significant number of individuals showed a meaningful (>5%) magnetoresistant effect, and a few of them were characterized in detail. Automated synthesis of such discrete material libraries, their analysis for different applications in material sciences, and the automated and reliable readout of the results make this class of libraries extremely useful, and in fact similar approaches are appearing more and more frequently in the literature.
162 COMBINATORIAL TECHNOLOGIES: BASIC PRINCIPLES
LnxMyCoOz
L33
prepared in duplicate as discrete individuals using materials science techniques
Ln= 2 lanthanides (La, Y)
M= 4 metals (Ba, Sr, Ca, Pb)
x/y= 16 different ratios
library size: 2x4x16=128 materials
composition of the 128 materials:
Ln= 4 different compositions: 0.62, 0.45, 0.33 and 0.27
M= 4 different compositions for each Ln and Ln value:
Ln=La, 0.62 M=1.38, 1.14, 0.89, 0.64
Ln=La, 0.45 M=1.00, 0.83, 0.65, 0.47
Ln=La, 0.33 M=0.73, 0.60, 0.47, 0.34
Ln=La, 0.27 M=0.61, 0.50, 0.39, 0.28
Ln=Y, 0.62 M=1.24, 1.00, 0.74, 0.50
Ln=Y, 0.45 M=0.90, 0.72, 0.54, 0.36
Ln=Y, 0.33 M=0.66, 0.52, 0.40, 0.26
Ln=Y, 0.27 M=0.55, 0.45, 0.33, 0.22
Figure 4.23 Structure of a materials science library L33.
REFERENCES
1.Spencer, R. W., Biotechnol. Bioeng. 61, 61–67 (1998).
2.Furka, A., Notarized document file number 36237/1982 , Dr. Judit Bokai, state notary public, June 15th, 1982, Budapest.
3.Geysen, H. M., Meloen, R. H. and Barteling, S. J., Proc. Natl. Acad. Sci. USA 81, 3998–4002 (1984).
4.Houghten, R. A., Proc. Natl. Acad. Sci. USA 82, 5131–5135 (1985).
5.Furka, A., Sebestyen, F., Asgedom, M. and Dibo, G., Int. J. Pept. Prot. Res. 37, 487–493 (1991).
6.Frank, R., Gueler, S., Krause, S. and Lindenmaier, W., Peptides 1990: Proceedings of Twenty First European Peptide Symposium , E. Giralt and D. Andrew (Eds.). ESCOM, Leiden, Netherlands, 1991, pp. 151–152.
7.Houghten, R., Pinilla, C., Blondelle, S. E., Appel, J. R., Dooley, C. T. and Cuervo, J. H., Nature 354, 84–86 (1991).
8.Lam, K. S., Salmon, S. E., Hersh, E. M., Hruby, V. J., Kazmierski, W. M. and Knapp, R. J., Nature 354, 82–84 (1991).
9.Fodor, S. P. A., Read, J. L., Pirrung, M. C., Stryer, L., Lu, A. T. and Solas, D., Science 251, 767–773 (1991).
10.Smith, G. P., Science 228, 1315–1317 (1985).
11.Parmley, S. F. and Smith, G. P., Gene 73, 305–318 (1988).
12.Devlin, J. J., Panganiban, L. C. and Devlin, P. E., Science 249, 404–406 (1990).
