
Garrett R.H., Grisham C.M. - Biochemistry (1999)(2nd ed.)(en)
.pdf
10.9 ● Ionophore Antibiotics |
321 |
stressed. To these ends, gap junctions are sensitive to membrane potentials, hormonal signals, pH changes, and intracellular calcium levels. Dramatic changes in pH or Ca2 concentration in a cell may be a sign of cellular damage or death. In order to protect neighboring cells from the propagation of such effects, gap junctions close in response to decreased pH or prolonged increases in intracellular Ca2 . Under normal conditions of intracellular Ca2 levels ( 10 7 M ), gap junctions are open and intercellular communication is maintained. When calcium levels rise to 10 5 M or higher, the junctions, sensing danger, rapidly close.
10.9 ● Ionophore Antibiotics
All of the transport systems examined thus far are relatively large proteins. Several small molecule toxins produced by microorganisms facilitate ion transport across membranes. Due to their relative simplicity, these molecules, the ionophore antibiotics, represent paradigms of the mobile carrier and pore or channel models for membrane transport. Mobile carriers are molecules that form complexes with particular ions and diffuse freely across a lipid membrane (Figure 10.38). Pores or channels, on the other hand, adopt a fixed orientation in a membrane, creating a hole that permits the transmembrane movement of ions. These pores or channels may be formed from monomeric or (more often) multimeric structures in the membrane.
Carriers and channels may be distinguished on the basis of their temperature dependence. Channels are comparatively insensitive to membrane phase transitions and show only a slight dependence of transport rate on temperature. Mobile carriers, on the other hand, function efficiently above a membrane phase transition, but only poorly below it. Consequently, mobile carrier systems often show dramatic increases in transport rate as the system is heated through its phase transition. Figure 10.39 displays the structures of several of these interesting molecules. As might be anticipated from the variety of structures represented here, these molecules associate with membranes and facilitate transport by different means.
● Schematic drawings of mobile carrier and channel ionophores. Carrier ionophores must move from one side of the membrane to the other, acquiring the transported species on one side and releasing it on the other side. Channel ionophores span the entire membrane.

322 Chapter 10 ● Membrane Transport
|
|
|
|
|
|
|
|
|
O |
|
|
CH3 |
O |
|
CH(CH3)2 |
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
(CH3)2CH |
|
O |
C |
C N |
|
|
|
|
|
|
|
|
|
|
||||||||||
|
|
|
|
|
C |
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
C |
H |
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
L-Lactate |
H |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
H |
|
H |
C |
|
|
CH(CH3)2 |
|
|
|
|
|
|
|||||||||
|
|
|
|
|
O |
|
NH D-Valine |
2 |
|
L-Valine |
O |
|
|
|
|
|
|
|
|||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
(CH3)2CH |
C |
|
|
1 |
|
|
|
|
3 |
|
HC |
|
|
O |
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
D-Hydroxyiso- C |
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
CH D-Hydroxyiso- |
|
|
|
valeric acid |
|
HN |
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
O |
valeric acid |
|
|
|
|
|
4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
HC |
|
CH(CH3)2 |
|
|
|
|
|
||||||||
|
|
|
|
O |
C |
|
|
12 |
|
|
|
|
|
|
D-Valine |
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
(CH3)2CH |
L-Valine |
|
|
|
|
|
|
|
|
5 |
|
|
C |
|
O |
|
|
|
|
|
|
|
||||
|
|
|
|
|
CH |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
11 |
|
|
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
NH |
|
|
|
|
|
|
|
|
L-Lactate |
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
O |
C |
L-Lactate |
|
|
|
|
|
|
6 |
HC |
CH3 |
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
CH |
|
10 |
|
|
|
|
|
L-Valine |
H |
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
CH3 |
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
O |
|
D-Valine |
D-Hydroxyiso- |
7 |
H |
N |
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
C |
|
9 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
H |
|
|
valeric acid |
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
O |
|
C |
|
H |
|
8 H |
O |
|
CH(CH3)2 |
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
(CH3)2CH |
|
N |
C |
|
C |
|
O |
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
O |
|
CH(CH3)2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
Valinomycin |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3 |
CH3 |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH |
3 |
|
O |
|
O |
O |
CH2 |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3 |
|||||
H3C |
|
|
CH3 |
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
HC |
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
O |
|
O |
|
|
O |
|
|
|
|
|
|
|
|
H3CO |
|
|
CH |
|
|
|
|
|
|
|
O |
|
||
O |
|
|
|
|
|
|
CH3 |
|
|
|
|
|
|
CH3 |
OH |
OH |
|
|
|||||||||||
|
|
|
|
CH3 |
|
|
|
|
|
|
|
|
H3C |
|
|
CH |
|
|
|
||||||||||
|
|
|
CH3 |
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
H2C |
O |
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
COO– |
|
|
|
|
|
|||||||
H3C |
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
CH |
|
|||||
O |
|
O |
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
HO |
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
O |
CH3 |
|
|
|
CH3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3 |
|
|
||
|
|
|
Nonactin |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Monensin |
|
|
|
|||
O |
H |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
HN |
LVal |
Gly |
LAla |
DLeu |
LAla |
DVal |
LVal |
DVal |
|
LTrp |
DLeu |
LTrp |
DLeu |
LTrp |
DLeu |
LTrp |
C |
|
|||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
NH |
|
|
|
|
|
|
|
|
|
|
|
|
Gramicidin A |
|
|
|
|
|
|
|
|
|
|
|
|
|
CH2 |
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
OH |
|
FIGURE 10.39 ● Structures of several ionophore antibiotics. Valinomycin consists of three repeats of a four-unit sequence. Because it contains both peptide and ester bonds, it is referred to as a depsipeptide.
10.9 ● Ionophore Antibiotics |
323 |
(a) |
(b) |
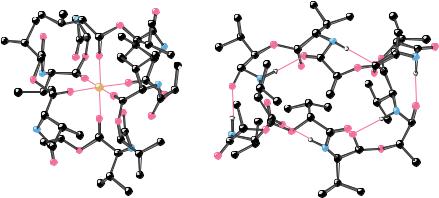
FIGURE 10.40 ● The structures of (a) the valinomycin-K complex and (b) uncomplexed valinomycin.
Valinomycin Is a Mobile Carrier Ionophore
Valinomycin (isolated from Streptomyces fulvissimus) is a cyclic structure containing 12 units made from four different residues. Two are amino acids (L-valine and D-valine); the other two residues, L-lactate and D-hydroxyiso- valerate, contribute ester linkages. Valinomycin is a depsipeptide, that is, a molecule with both peptide and ester bonds. (Considering the 12 units in the structure, valinomycin is called a dodecadepsipeptide.) Valinomycin consists of the 4-unit sequence (D-valine, L-lactate, L-valine, D-hydroxyisovalerate), repeated three times to form the cyclic structure in Figure 10.39. The structures of uncomplexed valinomycin and the K -valinomycin complex have been studied by X-ray crystallography (Figure 10.40). The structure places K at the center of the valinomycin ring, coordinated with the carbonyl oxygens of the 6 valines. The polar groups of the valinomycin structure are positioned toward the center of the ring, whereas the nonpolar groups (the methyl and isopropyl side chains) are directed outward from the ring. The hydrophobic exterior of valinomycin interacts favorably with low dielectric solvents and with the hydrophobic interiors of lipid bilayers. Moreover, the central carbonyl groups completely surround the K ion, shielding it from contact with nonpolar solvents or the hydrophobic membrane interior. As a result, the K -valinomycin complex freely diffuses across biological membranes and effects rapid, passive K transport (up to 10,000 K /sec) in the presence of K gradients.
Valinomycin displays a striking selectivity with respect to monovalent cation binding. It binds K and Rb tightly, but shows about a thousandfold lower affinity for Na and Li . The smaller ionic radii of Na and Li (compared to K and Rb ) may be responsible in part for the observed differences. However, another important difference between Na and K is shown in Table 10.5. The free energy of hydration for an ion is the stabilization achieved by hydrating that ion. The process of dehydration, a prerequisite to forming the ion-valinomycin complex, requires energy input. As shown in Table 10.5, considerably more energy is required to desolvate an Na ion than to desolvate a K ion. It is thus easier to form the K -valinomycin complex than to form the corresponding Na complex.
Other mobile carrier ionophores include monensin and nonactin (Figure 10.39). The unifying feature in all these structures is an inward orientation of polar groups (to coordinate the central ion) and outward orientation of non-







 amino acid Growing
amino acid Growing N
N