16-09-2014_09-00-17 / DNA structure
.doc
A nucleotide is composed of three parts: pentose, base and phosphate group. In DNA or RNA, a pentose is associated with only one phosphate group, but a cellular free nucleotide (such as ATP) may contain more than one phosphate group. If all phosphate groups are removed, a nucleotide becomes a nucleoside.
Cellular Nucleotides and Nucleosides
Nucleic Acid Chain
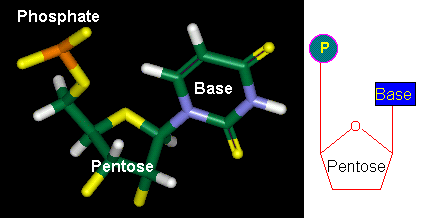
Figure 3-A-1. The general structure of nucleotides. Left: computer model. Right: a simplified representation.
Pentose

Figure 3-A-2. The chemical structure of pentose which contains five carbon atoms, labeled as C1' to C5'. The pentose is called ribose in RNA and deoxyribose in DNA, because the DNA's pentose lacks an oxygen atom at C2'. Recalling that RNA stands for "ribonucleic acid", and DNA for "deoxyribonucleic acid".
Bases
There are five different bases, each is denoted by a single letter as given in the parenthesis:
Adenine (A), Cytosine (C), Guanine (G), Thymine (T), and Uracil (U).
Among them,
A, C, G and T exist in DNA;
A, C, G and U exist in RNA.
Their chemical structures are shown in the following figure. A and G contain a pair of fused rings, classified as purines. C, T, and U contain only one ring, classified as pyrimidines.

Figure 3-A-3. Chemical structures of bases in DNA and RNA. The red dot is connected to the pentose.
We note that the chemical structure of uracil is simpler than thymine. This gives a good reason why RNA uses uracil, instead of thymine. However, why does DNA use thymine? The major requirement in designing DNA bases is that they must be able to form pairs. But adenine can pair with uracil as perfectly as with thymine (see Section B). Then, why DNA chooses a more complex base? This question had been puzzling researchers for many years, until they understood the DNA repairing mechanism (see Chapter 7 Section F).
In cells, a free nucleotide may contain one, two or three phosphate groups. The energy carrier ATP (adenosine triphosphate) has three phosphate groups; ADP (adenosine diphosphate) has two; AMP (adenosine monophosphate) has one. Their structures are shown in the following figure.

Figure 3-A-4. (a) A computer model of AMP. (b) Chemical structures of Adenosine, ADP and ATP. They differ in the number of phosphate groups.
If all phosphate groups are removed, a nucleotide becomes a nucleoside such as adenosine. The following table lists various nucleosides and nucleotides.
Table 3-A-1.
Cellular nucleosides and nucleotides.

NMP = nucleoside monophosphate NDP = nucleoside diphosphate NTP = nucleoside triphosphate dNMP = deoxynucleoside monophosphate dNDP = deoxynucleoside diphosphate dNTP = deoxynucleoside triphosphate
(nucleoside = ribonucleoside; deoxynucleoside = deoxyribonucleoside)
|
MoBio Contents |
The Nucleic Acid Chain |
|
Chapter 3 |
A | B | C | D | E | F | G | H |
|
|
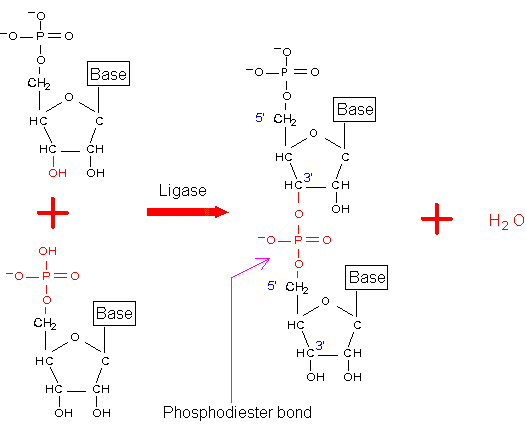
In a nucleic acid chain, two nucleotides are linked by a phosphodiester bond, which may be formed by the condensation reaction (Figure 3-A-5) similar to the formation of the peptide bond. In cells, such process has been found in the ligation between two nucleic acid fragments. However, the whole nucleic acid chain is usually synthesized by RNA polymerase or DNA polymerase.
Figure 3-A-5. Formation of the phosphodiester bond through the condensation reaction.
Like peptide chains, a nucleic acid chain also has orientation: its 5' end contains a free phosphate group and 3' end contains a free hydroxyl group. Synthesis of a nucleic acid chain always proceeds from 5' to 3'. Therefore, unless specified otherwise, the sequence of a nucleic acid chain is written from 5' to 3' (left to right).
Figure 3-A-6. A nucleic acid chain. Its 5' end contains a free phosphate group. The 3' end has a free hydroxyl group.
In DNA or RNA, a nucleic acid chain is also called a strand. A DNA molecule typically contains two strands whereas most RNA molecules contain a single strand. The length of a nucleic acid chain is represented by the number of bases. In the case of a double-stranded nucleic acid, bases are paired between two strands. Therefore, its length is given by the number of base pairs (bp). 1 kb = 1000 bases or bp; 1 Mb = 1 million bases or bp. Oligonucleotides refer to short nucleic acid chains (< 50 bases or bp) and polynucleotides have longer chains. Review Articles: Antisense Oligonucleotides: Basic Concepts and Mechanisms - Mol. Cancer Therap., 2002.
|
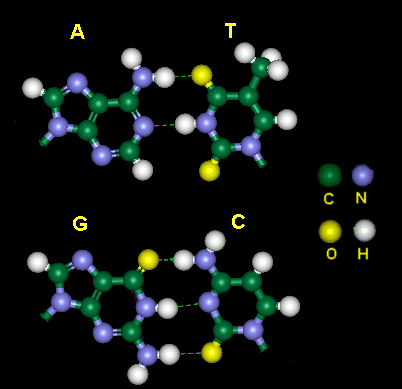
Figure 3-B-1. Computer model of base pairing in DNA. In a normal DNA molecule, adenine (A) is paired with thymine (T), guanine (G) is paired with cytosine (C). The uracil (U) of RNA can also pair with adenine (A), since U differs from T by only a methyl group located on the other side of hydrogen bonding.
A DNA molecule has two strands, held together by the hydrogen bonding between their bases. As shown in the above figure, adenine can form two hydrogen bonds with thymine; cytosine can form three hydrogen bonds with guanine. Although other base pairs [e.g., (G:T) and (C:T) ] may also form hydrogen bonds, their strengths are not as strong as (C:G) and (A:T) found in natural DNA molecules.
The following figure shows an example of base pairing between DNA's two strands.

Figure 3-B-2. Schematic drawing of DNA's two strands.
Due to the specific base pairing, DNA's two strands are complementary to each other. Hence, the nucleotide sequence of one strand determines the sequence of another strand. For example, in Figure 3-B-2, the sequence of the two strands can be written as
5' -ACT- 3'
3' -TGA- 5'
Note that they obey the (A:T) and (C:G) pairing rule. If we know the sequence of one strand, we can deduce the sequence of another strand. For this reason, a DNA database needs to store only the sequence of one strand. By convention, the sequence in a DNA database refers to the sequence of the 5' to 3' strand (left to right).
In a DNA molecule, the two strands are not parallel, but intertwined with each other. Each strand looks like a helix. The two strands form a "double helix" structure, which was first discovered by James D. Watson and Francis Crick in 1953. In this structure, also known as the B form, the helix makes a turn every 3.4 nm, and the distance between two neighboring base pairs is 0.34 nm. Hence, there are about 10 pairs per turn. The intertwined strands make two grooves of different widths, referred to as the major groove and the minor groove, which may facilitate binding with specific proteins.

Figure 3-B-3. The normal right-handed "double helix" structure of DNA, also known as the B form.
In a solution with higher salt concentrations or with alcohol added, the DNA structure may change to an A form, which is still right-handed, but every 2.3 nm makes a turn and there are 11 base pairs per turn.
Another DNA structure is called the Z form, because its bases seem to zigzag. Z DNA is left-handed. One turn spans 4.6 nm, comprising 12 base pairs. The DNA molecule with alternating G-C sequences in alcohol or high salt solution tends to have such structure.
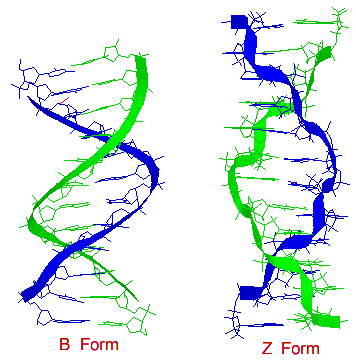
Figure 3-B-4. Comparison between B form and Z form.
Site of Interest: