
KAPLAN_USMLE_STEP_1_LECTURE_NOTES_2018_BIOCHEMISTRY_and_GENETICS
.pdf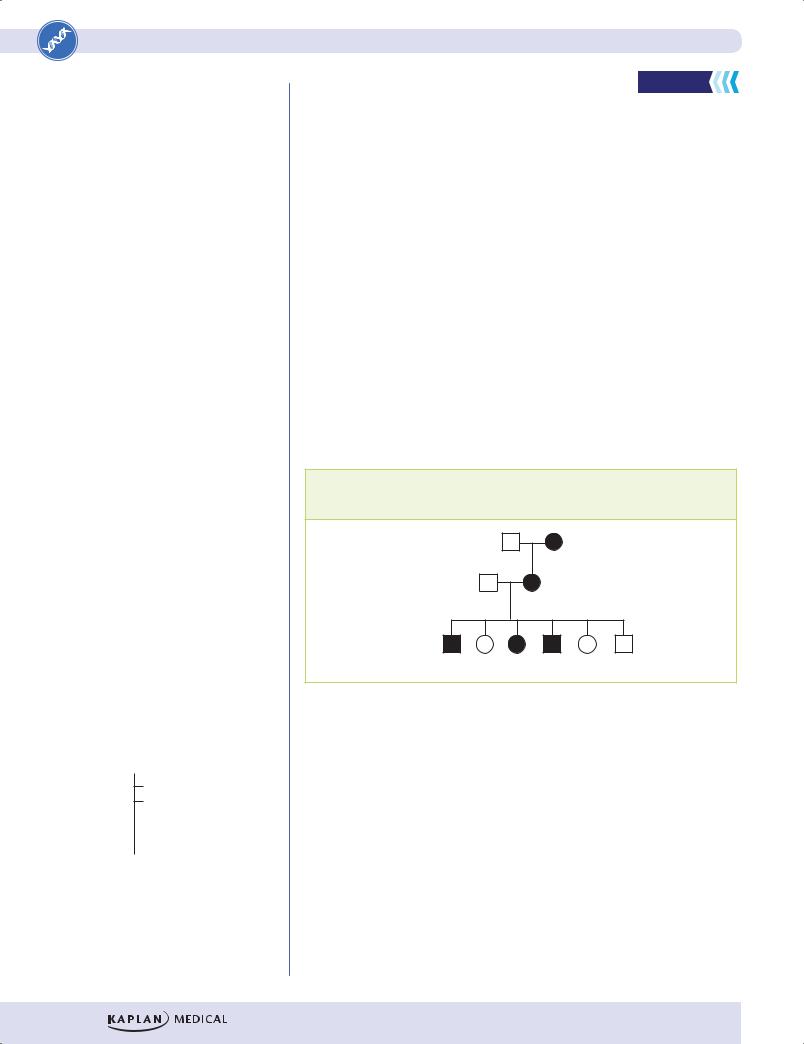



Chapter 5 ● Recombination Frequency
Review Questions
Select the ONE best answer.
1.A family with an autosomal dominant disorder is typed for a 2 allele marker, which is closely linked to the disease locus. Based on the individuals in Generation III, what is the recombination rate between the disease locus and the marker locus?
I
1,1 2,2
II
1,2 2,2
III
1,2 2,2 2,2 1,2
A.0
B.0.25
C.0.50
D.0.75
E.1.0
F.The marker is uninformative
2.A man who has alkaptonuria marries a woman who has hereditary sucrose intolerance. Both are autosomal recessive diseases and both map to 3q with a distance of 10 cM separating the two loci. What is the chance they will have a child with alkaptonuria and sucrose intolerance?
A.0%
B.12.5%
C.25%
D.50%
E.100%
387

Part II ● Medical Genetics
3.In a family study following an autosomal dominant trait through 3 generations, two loci are compared for their potential linkage to the disease locus. In the following 3-generation pedigree, shaded symbols indicate the presence of the disease phenotype, and the expression of ABO blood type and MN alleles are shown beneath each individual symbol.
AO OO
MN NN
AO OO
MN NN
AO AO OO AO OO OO AO
MN MN NN NN MN NN NN
Which of the following conclusions can be made about the linkage of the disease allele, ABO blood group locus, and MN locus?
A.The ABO and MN alleles are linked, but assort independently from the disease allele
B.The ABO, MN, and disease alleles all assort independently
C.The disease allele is linked to the ABO locus
D.The disease allele is linked to the ABO and MN loci
E.The disease allele is linked to the MN locus
388

Chapter 5 ● Recombination Frequency
Answers
1.Answer: A. In this pedigree, the disease allele is consistently transmitted with the 1 allele. There is no case in this small number of individuals where recombination between these two loci has occurred. Therefore, in Generation III, there is no recombination seen in any of the 4 individuals. Receiving the one allele always goes together with receiving the disease gene. Linked markers can be “uninformative” (choice E) in some pedigrees if, for example, the same alleles are expressed in all family members. In such a case, it would be impossible to determine any recombination frequency.
2.Answer: A. A child will inherit a gene for alkaptonuria from the father and the normal allele of this gene from the mother. Conversely, the child will inherit a gene for hereditary sucrose intolerance from the mother and a normal allele of this gene from the father. The child will therefore be a carrier for each disease but will not be affected with either one.
3.Answer: C. In this pedigree, the disease locus alleles are segregating with the ABO blood locus alleles. In each case, individuals who receive the A allele also receive the disease allele. The MN locus is not linked to the AO locus because individuals III-4, -5, and -7 are each recombinants between these loci. The MN locus is not linked to the disease allele because individuals III-4, -5, and -7 are each recombinants between these loci.
389


Genetic Diagnosis |
6 |
Learning Objectives
Understand concepts of direct and indirect genetic diagnosis
Understand concepts of allele-specific oligonucelotides and dot blots
GENETIC DIAGNOSIS
Once a gene is identified, the associated genetic disease in at-risk individuals can be diagnosed.
The goal of genetic diagnosis is to determine whether an at-risk individual has inherited a disease-causing gene. Genetic diagnosis can be distinguished into
2types:
•Direct diagnosis: the mutation itself is examined
•Indirect diagnosis: linked markers are used to infer whether the individual has inherited the chromosome segment containing the disease-causing mutation
Direct Diagnosis |
High-Yield |
|
PCR and allele-specific oligonucleotide (ASO) probes
ASO probes are short nucleotide sequences that bind specifically to a single allele of the gene. For example, the most common mutation causing hemochromatosis is the C282Y mutation that results from a G to A substitution in codon 282.
Codon: |
280 |
281 |
282 |
283 |
284 |
Normal HFE allele: |
TAT |
ACG |
TGC |
CAG |
GTG |
|
|
|
↑ |
|
|
C282Y allele: |
TAT |
ACG |
TAC |
CAG |
GTG |
|
|
|
↑ |
|
|
The ASO for the normal allele would have the sequence |
|
|
|||
3′ |
ATA |
TGC |
ACG |
GTC |
CAC 5′ |
The ASO for the C282Y allele would have the sequence |
|
|
|||
3′ |
ATA |
TGC |
ATG |
GTC |
CAC 5′ |
391

Part II ● Medical Genetics
The 2 ASOs could be used to probe the PCR-amplified material on a dot blot.
1 |
2 |
3 |
Normal ASO
C282Y ASO
Probe does not react with sample
 Probe reacts with sample
Probe reacts with sample
Figure II-6-1. Allele-Specific Oligonucleotide Probes in Hemochromatosis
The results show that individual 1 is homozygous for the normal HFE allele. Individual 2 is heterozygous for the normal and C282Y alleles. Individual 3 is homozygous for the C282Y allele. Only individual 3 would be expected to have symptoms. Note that this test merely determines genotype, and many considerations must be taken into account before predictions about phenotype could be made. Hemochromatosis has only about 15% penetrance, and in those who do have symptoms, variable expression is seen.
DNA chips
This approach involves embedding thousands of different oligonucleotides, representing various mutations and normal sequences, on a silicone chip. Patient DNA from specific regions is amplified by PCR, tagged with a fluorescent label, and exposed to the oligonucleotides on the chip. The sites of hybridization on the chip are recorded by a computer. This approach has the advantages of ready computerization and miniaturization (hundreds of thousands of oligonucleotides can be embedded on a single 2-cm2 chip).
392

Chapter 6 ● Genetic Diagnosis
Restriction fragment length polymorphism (RFLP) analysis of PCR products (RFLP-PCR)
Occasionally a mutation that creates a disease-producing allele also destroys (or creates in some instances) a restriction enzyme site, as illustrated by the following case:
A 14-year-old girl has been diagnosed with Gaucher disease (glucocerebrosidase A deficiency), an autosomal recessive disorder of sphingolipid catabolism. The mutation, T1448C, in this family also affects an HphI restriction site. PCR amplification of the area containing the mutation yields a 150-bp product. The PCR product from the normal allele of the gene is not cut by HphI. The PCR product of the mutant allele T1448C is cut by HphI to yield 114and 36-bp fragments. The PCR product(s) is visualized directly by gel electrophoresis. Based on the results shown below in Figure II-6-3 using this assay on DNA samples from this family, what is the most likely conclusion about sibling 2?
Affected |
Sibling 1 Sibling 2 |
Mother Father female |
150 bp
114 bp
36 bp
Figure II-6-2. PCR and RFLP for Gaucher Disease
(Ans.: Sibling 2 is also affected)
RFLP diagnosis of myotonic dystrophy
RFLP analysis is also useful in a few cases in which polymorphisms are too large to conveniently amplify with a PCR. One such case is myotonic dystrophy, in which the expanded sequence is within the gene region itself (a CTG in the 3′ untranslated region). This disease shows anticipation, and family members with a severe form of myotonic dystrophy may have several thousand copies of this repeat. As shown in Figure II-6-3, when EcoRI digests are analyzed by Southern blotting, a probe reveals 9- to 10-kb fragments in unaffected individuals. The size of the fragment can reach 20 kb in severely affected individuals.
393
