
- •Vary the numbers of specific enzymes made (regulation of gene expression)
- •Lac operon in e. Coli
- •Trp Operon - and example of a repressible operon
- •Enhancers
- •Silencers
- •E xample:
- •Split Genes
- •In general, introns tend to be much longer than exons. An average eukaryotic exon is only 140 nts long, but one human intron stretches for 480,000 nucleotides!
Enhancers
Some transcription factors ("Enhancer-binding protein") bind to regions of DNA that are thousands of base pairs away from the gene they control. Binding increases the rate of transcription of the gene.
Enhancers can be located upstream, downstream, or even within the gene they control.
How does the binding of a protein to an enhancer regulate the transcription of a gene thousands of base pairs away?
One possibility is that enhancer-binding proteins - in addition to their DNA-binding site, have sites that bind to transcription factors ("TF") assembled at the promoter of the gene.
This would draw the DNA into a loop (as shown in the figure).
Enhancers can work even if thier normal 5' to 3' orientation is flipped
Enhancers can work even if they are moved to a new location
Regulatory sequences with similar characteristics, but the opposite effect, exist. These are called silencers.
Silencers
Silencers are control regions of DNA that, like enhancers, may be located thousands of base pairs away from the gene they control. However, when transcription factors bind to them, expression of the gene they control is repressed.
Insulators
A problem:
As you can see above, enhancers can turn on promoters of genes located thousands of base pairs away. What is to prevent an enhancer from inappropriately binding to and activating the promoter of some other gene in the same region of the chromosome?
One answer: an insulator.
Insulators are
stretches of DNA (as few as 42 base pairs may do the trick)
located between the
enhancer(s) and promoter or
silencer(s) and promoter
of adjacent genes or clusters of adjacent genes.
Their function is to prevent a gene from being influenced by the activation (or repression) of its neighbors.
E xample:
The enhancer for the promoter of the gene for the delta chain of the gamma/delta T-cell receptor for antigen (TCR) is located close to the promoter for the alpha chain of the alpha/beta TCR (on chromosome 14 in humans). A T cell must choose between one or the other. There is an insulator between the alpha gene promoter and the delta gene promoter that ensures that activation of one does not spread over to the other.
All insulators discovered so far in vertebrates work only when bound by a protein designated CTCF ("CCCTC binding factor"; named for a nucleotide sequence found in all insulators). CTCF has 11 zinc fingers.
Another example: In mice (and humans), only the allele for insulin-like growth factor 2 (Igf2) inherited from one's father is active; that inherited from the mother is not - a phenomenon called imprinting.
The mechanism: the mother's allele has an insulator between the Igf2 promoter and enhancer. So does the father's allele, but in his case, the insulator has been methylated. CTCF can no longer bind to the insulator, and so the enhancer is now free to turn on the father's Igf2 promoter.
Post-Transcriptional Control
RNA Processing: pre-mRNA e mRNA
All the primary transcripts produced in the nucleus must undergo processing steps to produce functional RNA molecules for export to the cytosol. We shall confine ourselves to a view of the steps as they occur in the processing of pre-mRNA to mRNA.
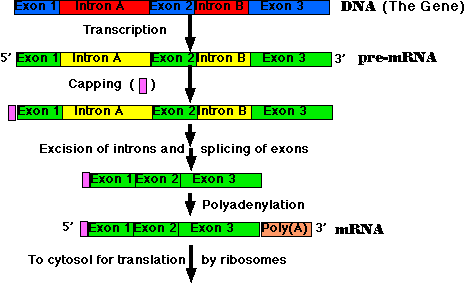
The steps:
Synthesis of the cap. This is a modified guanine (G) which is attached to the 5' end of the pre-mRNA as it emerges from RNA polymerase II (RNAP II). The cap protects the RNA from being degraded by enzymes that degrade RNA from the 5' end.
Step-by-step removal of introns present in the pre-mRNA and splicing of the remaining exons. This step is required because most eukaryotic genes are split. It takes place as the pre-mRNA continues to emerge from RNAP II.
Synthesis of the poly(A) tail. This is a stretch of adenine (A) nucleotides. When transcription is complete, the transcript is cut at a site (which may be hundreds of nucleotides before its end), and the poly(A) tail is attached to the exposed 3' end. This completes the mRNA molecule, which is now ready for export to the cytosol. (The remainder of the original transcript is degraded and the RNA polymerase leaves the DNA.)
