- •2) Objects and methods of animal biotechnology
- •3) Totipotent, multipotent, pluripotent animal cells
- •4.Allophenic animals. Genetic chimers
- •5)The principles of genetic cloning
- •6.Allophenic animals. Genetic chimers
- •8) Methods for introducing foreign dna into animal cells
- •9)Cryopreservation of reproductive and germ cells of animals and humans
- •11)The principles and methods of plant cells cultivation in vitro
- •12. The types of medium. Physiological means of compounds medium (as an example you can use the composition of Murashige-Skug medium)
- •14)Differentiation and dedifferentiation in plant cell culture. The obtaining callus mass and cultivation of callus tissue .
- •15)The influence of phytohormons on morphogenesis and regeneration in plant cells culture
- •16.The main path of morphogenesis processes in plant cells culture
- •18.The growth stages in suspension culture
- •20) The factors influenced on microclonal propagation in plant cell culture.
- •21) What is Biotechnology? Various definitions of “Biotechnology”. History of Biotechnology
- •22.Microbial Biotechnology: fundamentals of applied microbiology
- •24.Sterilization in Biotechnology: Methods and principles
- •26) Somaclonal and gametoclonal variation in plant cells culture.
- •27) Artificial seeds". Embryo culture in vitro
- •28. Culture of apical meristem cells
- •29)Cell reconstruction. Theoretical means of cell reconstruction
- •30.Basics of phytopathology. The main diagnostics methods of plant diseases
- •32) Main objects of animal biotechnology:
- •33) Morphological and functional features of gametes - eggs and sperm
- •34Hormonal regulation of mammalian reproduction
- •35)The history of investigations of the genetic transformation of animal cells
- •36.The principles of genetic engineering in animal biotechnology
- •53)Genetic engineering. Methods of genetic transformation
- •54. Methods of receiving plant materials without viruses
- •56) The vector systems used in the genetic engineering
- •57) Methods of genetic engineering: agrobacterial genetic transformation
- •58)Methods of genetic engineering: bioballistics methods
- •60.Apply cell technology and cryopreservation technology for safe gene bank
- •62) Methods of producing chimeras
- •63) Collection and cultivation of oocytes in vivo and in vitro
- •64 Collection and cultivation of embryos in vivo and in vitro
- •66.Fertilization of oocytes in vitro, environment and conditions
- •68) Draw a diagram of the structure of plasmid pBr322
- •69) Draw a diagram of an experiment in genetic engineering (design recDna) and give a description of the main stages
- •70)Describe the calcium-phosphate method for introducing foreign dna into mammalian cells.
- •72 Methods of cryopreservation of sperm and oocytes of mammals
- •74) Modes of freezing and thawing of gametes and embryos
- •75) Methods of artificial fertilization: gamete insemination fallopian tube (gift), zygosity insemination fallopian tubes (zift).
- •76) Stem cells and prospects for their use in practice
- •78.Technical equipment of experiments on artificial insemination
- •80) Methods of animal cloning, reproductive and therapeutic cloning
- •81) Microorganisms in water and wastewater treatment
- •82 Microbial fermentations in food products
- •84.Bacterial examination of water and standard water analysis
- •86) Use of e.Coli for the biotechnological production
- •87) Microbes in milk and dairy products
- •88) What is the benefit of microorganisms in industry
- •90. Algae, their applications
68) Draw a diagram of the structure of plasmid pBr322
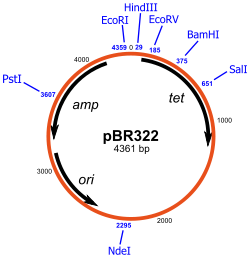
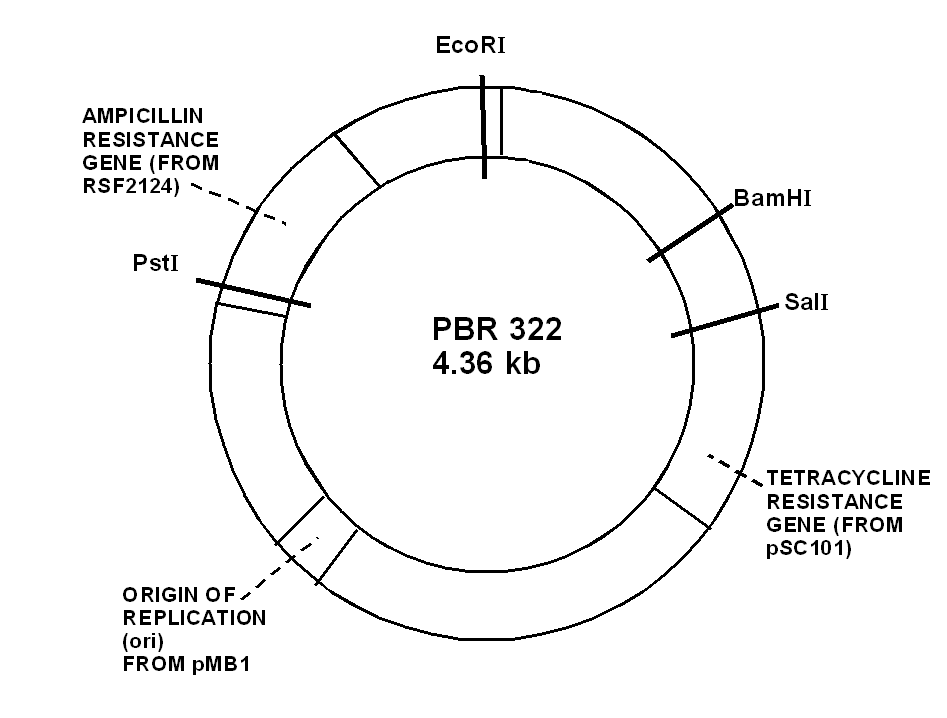
69) Draw a diagram of an experiment in genetic engineering (design recDna) and give a description of the main stages
Molecular cloning is the laboratory process used to create recombinant DNA. It is one of two widely used methods (along withpolymerase chain reaction, abbr. PCR) used to direct the replication of any specific DNA sequence chosen by the experimentalist. The fundamental difference between the two methods is that molecular cloning involves replication of the DNA within a living cell, while PCR replicates DNA in the test tube, free of living cells.
Formation of recombinant DNA requires a cloning vector, a DNA molecule that will replicate within a living cell. Vectors are generally derived from plasmids or viruses, and represent relatively small segments of DNA that contain necessary genetic signals for replication, as well as additional elements for convenience in inserting foreign DNA, identifying cells that contain recombinant DNA, and, where appropriate, expressing the foreign DNA. The choice of vector for molecular cloning depends on the choice of host organism, the size of the DNA to be cloned, and whether and how the foreign DNA is to be expressed.[5] The DNA segments can be combined by using a variety of methods, such as restriction enzyme/ligase cloning or Gibson assembly.
In standard cloning protocols, the cloning of any DNA fragment essentially involves seven steps: (1) Choice of host organism and cloning vector, (2) Preparation of vector DNA, (3) Preparation of DNA to be cloned, (4) Creation of recombinant DNA, (5) Introduction of recombinant DNA into the host organism, (6) Selection of organisms containing recombinant DNA, (7) Screening for clones with desired DNA inserts and biological properties.
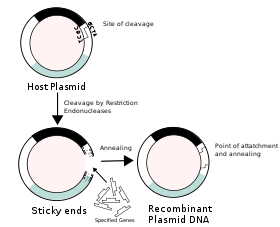
Construction of recombinant DNA, in which a foreign DNA fragment is inserted into a plasmid vector. In this example, the gene indicated by the white color is inactivated upon insertion of the foreign DNA fragment.
70)Describe the calcium-phosphate method for introducing foreign dna into mammalian cells.
Calcium phosphate transfection, initially described in the early 1960s, was refined and systematically improved to result in a standard protocol which has changed little since the early 1970s. In many cases it remains the system of choice for transferring plasmid DNA into a variety of cell cultures and packaging cell lines. It is particularly important in the production of recombinant viral vectors.
Basics of the technique. The technique relies on precipitates of plasmid DNA formed by its interaction with calcium ions. It is a very inexpensive and simple technique to perform. Plasmid DNA is mixed in a solution of calcium chloride, and then is added to a phosphate- buffered solution. Over a period of 20 minutes a fine precipitate forms in the solution, and this solution is then added directly to the cells in culture. Transfer efficiency, the number of cells which express the desired gene, is usually quite limited and only reaches levels greater than 10% in a few specific cell lines. In many cases, the level is less than 1%. Transfection efficiencies can be improved in some cell lines by 'shocking' the cells with DMSO or glycerol. Cells can be either transiently or stably transfected using this technique. Levels of stable transfectants can be improved using bis-hydroxyethylaminoethansulfonate (BES). The DNA precipitates are thought to enter the cell by endocytosis. Although this technique has minimal cellular toxicity, and is both simple and inexpensive, the low level of transgene expression prompted development of other techniques. Little or no transgene expression has been observed in vivo following calcium phosphate transfection.
DEAE-dextran is another compound that interacts and delivers DNA into cells. It is more reproducible than calcium phosphate transfection but only works in a select few cell lines. Also, DEAE-dextran-mediated gene transfer results only in transient transfections. The exact mechanism of DNA uptake mediated by this technique is not understood.
71 |
)Describe the method of microinjection of DNA in the germ cells of animals |
Microinjection is an effective method for creating transgenic animals, for RNAi of selected genes, and for introducing various types of molecules directly to cells. For DNA transformation, the easiest approach is to inject DNAs into the distal arm of the gonad. The distal germ line of C. elegans contains a central core of cytoplasm that is shared by many germ cell nuclei .Therefore, DNAs injected here can be delivered to many progeny. This approach usually leads to the formation of large extrachromosomal DNA arrays . Microinjection directly into oocyte nuclei can induce chromosomal integration of transgenes, but this technique is relatively difficult to do . For RNAi experiments, most progeny of injected animals can be affected by simply injecting dsRNA into a single gonad or intestinal cell because of a very efficient RNA transport system . While RNAi by feeding is best for high throughput experiments, RNAi by microinjection is more effective for at least some genes .
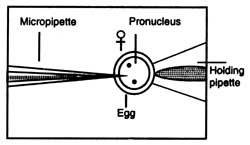
Microinjection Microinjection is a technique of delivering foreign DNA into a living cell (a cell, egg, oocyte, embryos of animals) through a glass micropipette. One end of a glass micropipette is heated until the glass becomes somewhat liquified. It is quickly stretched which forms a very fine tip at the heated end. The tip of the pipette attains to about 0.5 mm diameter which resembles an injection needle. The process of delivering foreign DNA is done under a powerful microscope. Cells to be microinjected are placed in a container (Fig. 4.15). A holding pipette is placed in the field of view of the microscope. The holding pipette holds a target cell at the tip when gently sucked. The tip of the micropipette is injected through the membrane of the cell. Contents of the needle are delivered into the cytoplasm and the empty needle is taken out.
|
|
|
|
Fig. 4.15. A method of microinjection of DNA preparation in egg. |
|
Xenopus oocytes have been widely used for the study of transcription by microinjection because oocytes contain between 6,000 and 100,000 or more RNA polymerase molecules than somatic cells. Microinjection is technically easy because of large size of oocytes. Some of the endogenous pattern of gene regulation during development has been characterized (Wickens and Laskey, 1981). The injected DNA integrates randomly with nuclear DNA and its expression could be possible only when the foreign DNA is attached to a suitable promoter sequence.