
- •Иерархическая классификация гликозил-гидролаз: современное состояние и перспективы развития
- •Классификация Ферментов (IUBMB)
- •Sequence Based Classification of Glycoside Hydrolases
- •Sequence Based Classification of Glycoside Hydrolases
- •Sequence Based Classification of Glycoside Hydrolases
- •Оптическая конфигурация гликозидной связи (аксиальная или экваториальная) и механизм её гидролиза (с сохранением
- •Объединение семейств гликозидаз в кланы
- •Структура и гидролиз раффинозы
- •Утилизация сахарозы бактериями: типы -фруктозидаз
- •Подсемейства семейства GH32
- •Филогенетическое древо семейства GH32
- •Подсемейства семейства GH68
- •Множественное выравнивание последовательностей белков, представляющих семейства -фруктозидазного суперсемейства
- •Family
- •Структура -фруктозидазного (фуранозидазного) суперсемейства
- •Пространственная структура белков семейства GH32
- •Семейства гликозидаз, содержащие α-галактозидазы
- •Family GH27
- •Пространственная структура белков семейства GH27 гликозидаз
- •Family GH36
- •Четыре консервативные участка в белках α-галактозидазного суперсемейства, содержащие остатки Asp
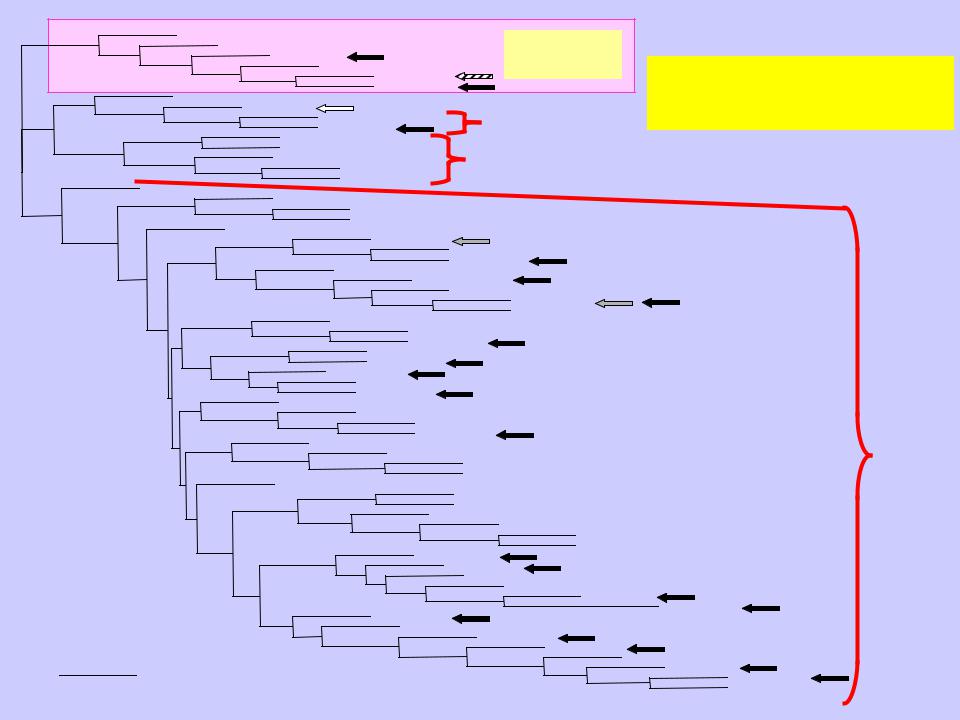
- •Филогенетическое древо α-галактозидазного суперсемейства
- •Family
- •Rigden DJ. Iterative database searches demonstrate that glycoside hydrolase families 27, 31, 36,
- •(β/α)8-barrel-type retaining α D-glycopyranosidase families
- •Родственные связи (β/α)8 гликозидаз (Nagano et al., 2001)
- •Объединение семейств гликозидаз в кланы
- •Объединение семейств гликозидаз в кланы
- •Иерархическая классификация гликозил-гидролаз
- •Персональные гранты, поддержавшие исследования в области анализа аминокислотных последовательностей

Family GH36
Subfamily GH36A:
–bacterial α-galactosidases: Azotobacter, Bacillus, Geobacillus, Bifidobacterium, Carnobacterium, Clostridium, Enterococcus, Erwinia, Escherichia, Geobacillus, Klebsiella, Lactobacillus, Lactococcus, Leuconostoc, Novosphingobium, Oenococcus, Pediococcus, Ruminococcus, Streptococcus, Streptomyces, Thermoanaerobacter, Treponema, Vibrio, and Yersinia
–fungal α-galactosidases: Absidia, Aspergillus, Gibberella, Penicillium, and Trichoderma
Subfamily GH36B:
– bacterial α-galactosidases: Burkholderia, Leptospira, Streptomyces, Thermotoga, Thermus, and Vibrio
Subfamily GH36C:
–plant alkaline α-galactosidases or seed imbibition proteins: Arabidopsis, Brassica, Cicer, Cucumis, Hordeum, Lycopersicon, Malus, Oryza, and Persea
–plant α-galactosyltransferases: Alonsoa, Arabidopsis, Cucumis, Medicago, Oryza, Pisum, Stachys, and Vigna
–fungal ORFs: Aspergillus, Coccidioides, Cryptococcus, Gibberella, Magnaporthe, Neurospora, and Ustilago
–ORF from Toxoplasma gondii
–α-galactosidase from Bifidobacterium breve
–bacterial ORFs: Bifidobacterium longum, Bacteroides fragilis, and Bacteroides thetaiotaomicron
–ORFs from Sulfolobus solfataricus and Sulfolobus tokodaii
Subfamily GH36D:
–α-N-acetylgalactosaminidase from Clostridium perfringens
–bacterial ORFs: Aeromonas, Clostridium, Escherichia, Photorhabdus, and Streptococcus

Четыре консервативные участка в белках α-галактозидазного суперсемейства, содержащие остатки Asp
MEL1_YEAST |
65 |
GYKYIILDDCWS |
141 |
NRVDYLKYDNCY |
204 |
WRMSGDV |
258 |
WNDLDNL |
AGAL_PHAVU |
106 |
GYQYINIDDCWG |
184 |
WGIDYLKYDNCE |
242 |
WRTTGDI |
274 |
WNDPDML |
NAGA_HUMAN |
71 |
GYTYLNIDDCWI |
148 |
WKVDMLKLDGCF |
212 |
WRNYDDI |
248 |
WNDPDML |
AGL1_BIFLO |
48 |
GWDTLVIDIDWY |
178 |
WGLDFLKVDDMQ |
240 |
WRISDDL |
272 |
WADADMV |
AGL3_HYPJE |
234 |
GYDLCSLDSGWQ |
339 |
WGVDMLKLDFLT |
407 |
MRTDQDL |
451 |
YPDMDAL |
IMD_ARTGO |
100 |
GYDIACTD-GWI |
219 |
LGVPYLRIDFLS |
290 |
VRINADA |
338 |
ILDGDFM |
AGAL_LACPL |
367 |
GIEMFVLDDGWF |
478 |
VPIDYIKWDMNR |
551 |
QSWPSDN |
584 |
GTSPDEL |
AGL2_HYPJE |
375 |
GIKLFVLDDGWF |
496 |
ASISYVKWDNNR |
561 |
HIWTSDD |
594 |
SAVPNGQ |
AGL7_ASPFU |
413 |
GAGYFVIDAGWY |
524 |
YGVGYFKFDYNI |
595 |
LQSSSDQ |
626 |
WAYPQPA |
AGAL_THEMA |
213 |
PFEVFQIDDAYE |
319 |
MGYRYFKIDFLF |
382 |
MRIGPDT |
423 |
LNDPDCL |
AGAL_VIBPA |
227 |
DLEWVLLDDGYQ |
341 |
WGVELFKLDANY |
404 |
MRVSDDV |
436 |
QIDPDCA |
AGL3_STRCO |
329 |
GLKWAVLDDGWQ |
438 |
WGYEGLKIDGQH |
512 |
QYPSSDP |
540 |
SYSGDHV |
AGAL_SULSO |
256 |
RLNWVIIDDGWQ |
360 |
RDFDLVKVDNQW |
421 |
MRNSIDY |
454 |
YPDYDMF |
AGAL_BIFBR |
339 |
PVSWVLIDDGWS |
458 |
AGVDFVKVDSQS |
523 |
TRTSDDF |
556 |
HCDWDMF |
GALT_VIGAN |
251 |
APRFVVIDDGWQ |
476 |
TGVTGVKIDVIH |
544 |
GRVGDDF |
585 |
QPDWDMF |
NAGA_CLOPE |
253 |
TLDAFVVDDGWA |
355 |
YDISYWKIDGML |
429 |
IQTSQDV |
|
... |
ORF1_CLOPE |
515 |
PIDSYVVDDGWH |
634 |
FDIDYWKLDGFA |
715 |
IQNSQDT |
|
... |
ORF1_ECOLI |
297 |
ALDAFLLDDGWD |
393 |
EHITSFKLDGMG |
458 |
WRQGDDI |
|
... |
ORF2_CLOPE |
112 |
PKGIIMIDDGWS |
220 |
YGVDGFKFDAGD |
286 |
HSWEYNG |
344 |
ALMPMMQ |
XYLQ_LACPE |
297 |
PLDVFHFDCFWQ |
406 |
MGVDSFKTDFGE |
474 |
IQYTGAA |
534 |
LLSSHSR |
XYLS_SULSO |
250 |
PLDVIVLDWRYW |
345 |
LGIDAYWLDASE |
423 |
ISWSGDV |
483 |
TFCPILR |
LYAG_HUMAN |
397 |
PLDVQWNDLDYM |
510 |
VPFDGMWIDMNE |
611 |
GHWTGDV |
666 |
AFYPFMR |
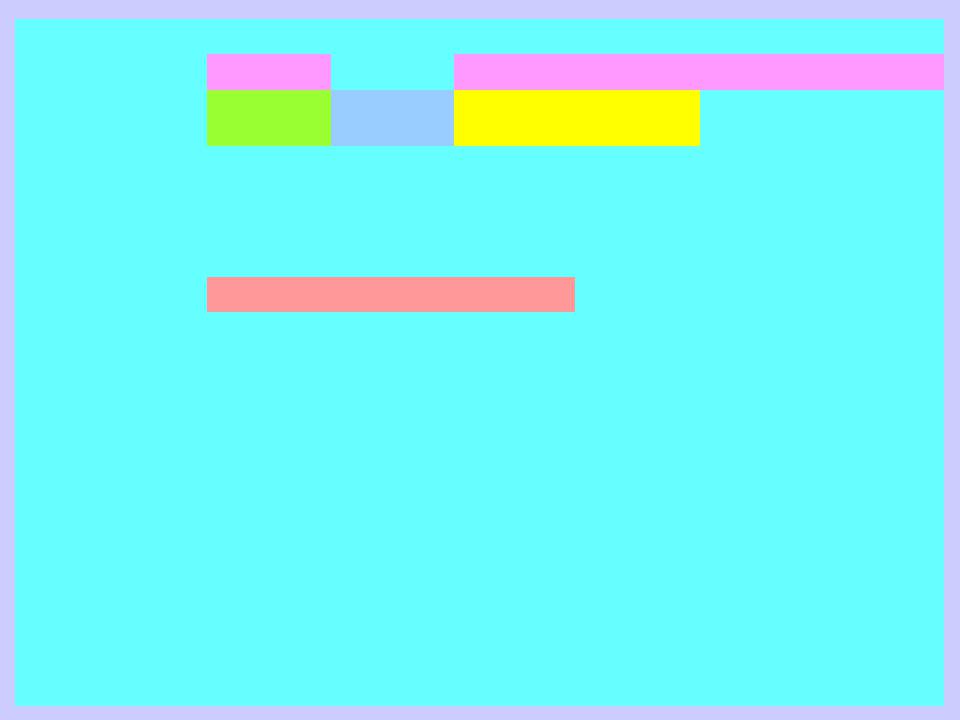
GH27
GH36A
GH36B
GH36C
GH36D
GH31
Нуклеофил |
Донор H+ |

Филогенетическое древо α-галактозидазного суперсемейства
|
|
XYLS SULSO |
89 |
|
|
|
|
|
|
|
|
|
|
|
||
|
AGL2 BACTQ |
|
|
|
|
|
|
|
|
|
|
GH27 |
||||
|
AGLU ACIAC |
38 |
59 |
|
|
|
|
|
|
|
|
|
|
|
||
SUIS HUMANc |
|
|
|
|
|
|
|
|
|
AGAL PORGI |
|
|||||
SUIS HUMANn |
55 |
|
72 |
|
|
86 |
|
|
|
|
16 |
|
AGAL SACER |
|
|
|
LYAG HUMAN |
|
|
|
|
|
|
|
|
|
|
|
AGAL PSEFL |
||||
|
|
|
|
|
|
|
|
|
|
|
97 |
|||||
|
XYLQ LACPE |
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
9 |
|
AGAL MICDE |
||||||
|
|
|
77 |
|
|
|
|
|
|
|||||||
|
ORF1 THEMA |
|
|
|
|
|
|
|
MEL2 ARATH |
|||||||
|
40 |
|
69 |
|
|
|
|
61 |
|
|||||||
|
ORF1 BACHA |
70 |
|
|
|
|
|
|
|
100 |
|
AGAL CYATE |
||||
YICI ECOLI |
|
|
|
|
|
|
|
|
|
|
|
|
AGAL PHAVU |
|||
|
42 |
|
|
|
|
|
5 |
|
|
AGL1 STRCO |
|
|||||
ORF1 CLOAC |
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
||||||
GH31 |
|
|
|
ORF1 CHLAU |
|
43 |
|
7 |
5 |
16 |
|
AGL2 ASPFU |
|
|
||
|
|
ORF2 CLOPE |
36 |
25 |
|
6 |
67 |
|
AGAL FIBSU |
|||||||
|
ORF1 MOUSE |
|
80 |
|
|
|
|
AGAL CLOJO |
||||||||
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
ORF1 DROME |
|
|
|
|
|
|
|
33 |
AGLB ASPNG |
|
||||
|
|
|
ORF1 AERHY |
93 |
|
|
|
|
|
MEL1 YEAST |
||||||
GH36D |
|
ORF1 ECOLI |
|
|
|
21 |
|
27 |
|
MELA PHACH |
||||||
|
92 |
98 |
30 |
|
|
|
|
|||||||||
ORF1 STRPN |
|
|
|
|
|
|
98 |
NAGA ACRSP |
||||||||
|
|
|
NAGA CLOPE |
|
|
|
|
|
|
|
|
|
|
AGLA ASPNG |
||
|
|
ORF1 CLOPE |
|
39 |
|
|
|
84 |
|
|
48 |
|
MEL1 CAEEL |
|||
|
|
|
AGL3 STRCO |
|
|
|
|
46 |
|
|||||||
|
|
|
|
|
|
|
|
|
|
80 |
|
MEL1 DROME |
||||
|
|
|
|
AGL2 STRCO |
|
24 |
|
7 |
|
|
|
|
NAGA HUMAN |
|||
GH36B |
|
AGAL THETH |
|
|
57 |
|
|
|
|
|
|
56 |
AGAL HUMAN |
|||
|
|
72 |
|
|
|
|
100 |
|
|
|||||||
|
AGAL THEMA |
|
|
|
|
|
|
AGL2 BACFR |
||||||||
|
|
|
AGAL THET2 |
77 |
37 |
|
|
|
|
AGL3 BACFR |
|
|
||||
|
|
|
|
|
20 |
|
12 |
|
|
|||||||
|
|
|
AGAL LEPIN |
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
45 |
|
|
|
|
|
AGL1 BACFR |
||||
|
|
|
|
AGAL VIBCH |
99 |
|
|
|
AGL3 HYPJE |
|
|
|||||
|
|
|
|
AGAL VIBPA |
|
|
72 |
|
|
IMD ARTGO |
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
AGA2 PEDPE |
|
|
|
|
39 |
|
|
94 |
|
MEL4 ARATH |
||
|
|
|
AGA1 PEDPE |
|
|
|
|
|
|
|
|
|
MEL5 ORYSA |
|||
|
|
|
|
|
|
|
|
|
|
49 |
|
|
|
|||
|
AGAL LACPL |
39 |
49 |
|
|
|
|
|
|
47 |
AGL1 BIFLO |
|||||
|
AGAL STRMU |
40 |
|
|
|
|
|
|
|
62 |
|
AGAL BACHA |
||||
|
|
|
86 |
|
|
|
|
|
|
|||||||
|
AGL2 RUMAL |
54 |
|
|
|
|
|
|
|
|
|
AGL1 RUMAL |
||||
|
|
|
|
|
|
|
|
AGAL SULTO |
|
|||||||
|
AGL5 BACFR |
|
94 |
|
|
21 |
|
|
|
93 |
|
GH36C |
||||
|
|
|
|
79 |
39 |
|
89 |
|
AGAL BIFBR |
|
||||||
|
AGLC ASPNG |
|
|
|
76 |
|
||||||||||
|
AGL6 BACFR |
AGL6 ASPFU |
|
|
|
|
|
|
|
|
AGAL SULSO |
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
AGL4 BACFR |
|
|
|||
|
AGL2 HYPJE |
69 |
|
|
|
65 |
36 |
|
|
|
|
ORF2 ARATH |
||||
|
|
|
|
|
|
|
45 |
|
||||||||
|
|
|
AGAL ABSCO |
|
|
|
|
|
|
|
|
|
STAS PISSA |
|||
|
|
|
|
31 |
|
|
|
|
|
|
67 |
|
||||
|
|
|
AGL2 BIFLO |
51 |
|
|
|
|
|
94 |
|
|
GALT VIGAN |
|||
|
|
|
RAFA ECOLI |
|
|
|
|
|
|
|
|
|
||||
GH36A |
|
|
|
|
|
|
|
|
58 |
ORF1 ARATH |
||||||
|
|
AGL7 ASPFU |
|
|
|
|
|
|
|
|
SIP HORVU |
|||||
|
|
|
|
AGL3 RUMAL |
|
99 |
|
|
|
|
53 |
|
SIP CICAR |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
AGAL SULSO |
|
|
ORF1 NEUCR |
|
GH36C |
|
|
||||
995 |
670 |
AGL5 BACTH |
|
|
|
|||||||
|
519 |
|
|
AGAL BIFBR |
AGA1 CUCME |
|
|
|
Филогенетическое древо |
|||
|
|
623 |
|
713 |
|
|
|
|
||||
|
|
|
|
|
STAS STAAF |
|
|
|
|
|
||
532 |
AGL4 STRAV |
|
|
AGL8 ASPNI |
|
|
|
|
|
семейства GH27 |
||
879 |
IMD ARTGO |
|
27c |
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
||
410 |
|
983 |
|
|
AGL3 HYPJE |
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
||||
|
999 |
|
|
MEL4 ARATH |
|
|
27b |
|
|
|
|
|
|
|
|
MEL5 ORYSA |
|
|
|
|
|
|
|||
|
903 |
|
|
|
|
|
|
|
|
|||
|
|
|
AGL1 BIFLO AGAL BACHA |
|
|
|
|
|||||
|
905 |
|
|
|
|
|
|
|||||
|
|
|
862 |
AGL1 RUMAL |
|
|
|
|
|
|||
|
AGL3 STRAV |
|
|
|
|
|
|
|
|
|||
|
|
|
AGL3 BACFR |
|
|
|
|
|
|
|
||
|
988 |
|
|
AGL4 BACTH |
|
|
|
|
|
|||
513 |
|
|
996 |
|
|
|
|
|
|
|||
|
AGL5 BACTH |
|
AGL3 BACTH |
|
|
|
|
|
||||
|
721 |
|
NAGA ACRSP |
|
|
|
|
|
||||
|
|
|
993 |
|
AGL4 ASPFU |
|
|
|
|
|||
|
616 |
|
|
999 |
|
|
|
|
|
|||
|
|
|
AGL2 DICDI |
|
AGLA ASPNG |
|
|
|
|
|||
|
372 |
|
525 |
AGAL HUMAN |
|
|
|
|
||||
|
|
996 |
|
|
|
|
|
|||||
|
|
|
|
|
479 |
|
MEL1 CAEEL |
|
|
|
|
|
|
|
|
|
|
|
794 |
NAGA HUMAN |
|
|
|||
|
265 |
|
|
|
AGL1 STRAV |
|
MEL1 DROME |
|
|
|||
|
906 |
|
AGL2 STRAV |
|
|
|
|
|||||
|
124 |
|
|
|
1000 |
|
AGL1 STRCO |
|
|
|
|
|
|
|
|
998 |
|
AGAL MICDE |
|
|
|
|
|
||
|
378 |
|
|
AGAL PSEFL |
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
||||
|
|
|
AGAL SACER |
|
|
|
|
|
|
|||
|
|
479 |
|
|
|
|
|
|
|
|||
|
|
376 |
|
AGAL FIBSU |
|
|
|
|
|
|||
|
57 |
|
|
|
AGAL CLOJO |
|
|
|
|
|
||
|
|
|
AGL1 DICDI |
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|||
|
265 |
|
|
MEL6 ORYSA |
|
|
|
|
|
|||
|
|
|
990 |
|
|
|
|
|
|
|||
|
|
|
|
767 |
MEL2 ORYSA |
|
|
|
|
|||
|
|
|
|
|
MEL1 ORYSA |
|
|
|
|
|||
|
103 |
|
|
AGAL PORGI |
|
|
|
27a |
||||
|
580 |
|
|
|
|
|
|
|||||
|
|
|
1000 |
AGL1 BACFR |
|
|
|
|||||
|
|
|
|
973 |
AGL1 BACTH |
|
|
|
||||
|
76 |
|
|
AGL2 BACTH |
|
|
AGL2 BACFR |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
AGL2 ASPNI |
|
|
|
|
||
|
|
|
|
|
|
1000 |
|
|
|
|
|
|
|
|
|
|
827 |
|
|
AGL2 ASPFU |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||
|
143 |
|
|
|
689 |
AGL1 USTMA |
|
|
|
|
||
|
|
|
|
|
|
885 |
AGL2 GIBZE |
AGL3 ASPFU |
|
|||
|
|
|
|
|
|
|
1000 |
|
|
|||
|
|
|
|
|
|
|
|
|
AGL3 ASPNI |
|
||
|
|
466 |
|
|
|
|
MELA PHACH |
|
|
|||
|
|
|
|
975 |
|
|
|
|
|
|||
|
|
|
|
|
386 |
MEL1 UMBVI |
|
|
|
|
||
|
|
|
|
|
|
|
MEL1 SCHPO |
|
|
|
|
|
|
|
|
|
|
|
250 |
|
|
|
|
||
|
|
|
|
|
|
AGL1 ASPFU |
|
|
|
|||
|
|
|
342 |
|
|
515 |
MEL5 YEAST |
|
||||
|
|
|
|
|
|
|
|
1000 |
|
|
MEL1 SACKL |
|
|
|
|
|
|
|
MEL2 UMBVI |
|
|
|
|||
|
|
|
|
|
374 |
|
|
|
|
|
||
|
|
|
|
421 |
|
AGL1 GIBZE AGL1 HYPJE |
|
|
|
|||
1000 |
|
|
|
|
|
|
994 |
866 |
AGL1 PENPU |
AGL5 ASPNI |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
987 |
554 |
|
AGL1 PENSIAGLB ASPNG |
|
|
|
|
|
|
|
|
|
|
814 |
||
|
|
|
|
|
|
|
|
|
|
|
AGL5 ASPFU |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Family |
GH27 |
GH31 |
GH36A |
GH36B |
GH36C |
GH36D |
|
|
|
|
|
|
|
Clan |
GH-D |
None |
GH-D |
GH-D |
GH-D |
GH-D |
|
|
|
|
|
|
|
COG / KOG |
KOG2366 |
COG1501 |
COG3345 |
COG3345 |
None |
None |
|
|
KOG1065 |
|
|
|
|
Known enzymatic |
EC 2.4.1.x |
EC 3.2.1.3 |
EC 3.2.1.22 |
EC 3.2.1.22 |
EC 2.4.1.67 |
EC 3.2.1.49 |
activities |
EC 3.2.1.22 |
EC 3.2.1.10 |
|
|
EC 2.4.1.82 |
|
|
EC 3.2.1.49 |
EC 3.2.1.20 |
|
|
EC 3.2.1.22 |
|
|
EC 3.2.1.94 |
EC 3.2.1.48 |
|
|
|
|
|
|
EC 4.2.2.13 |
|
|
|
|
|
|
|
|
|
|
|
Molecular mechanism |
Retaining |
Retaining |
Retaining |
Not known |
Not known |
Not known |
|
|
|
|
|
|
|
Origin |
Eukaryota: |
Eukaryota: |
Eukaryota: |
Eubacteria: |
Eukaryota: |
Eubacteria: |
|
Alveolata |
Alveolata |
Fungi |
Actinobacteria |
Alveolata |
Firmicutes |
|
Fungi |
Fungi |
Eubacteria: |
Proteobacteria |
Fungi |
Proteobacteria |
|
Metazoa |
Metazoa |
Actinobacteria |
Spirochaetes |
Viridiplantae |
|
|
Mycetozoa |
Mycetozoa |
Bacteroidetes |
Thermotogales |
Eubacteria: |
|
|
Viridiplantae |
Rhodophyta |
Firmicutes |
Thermus |
Actinobacteria |
|
|
Eubacteria: |
Viridiplantae |
Proteobacteria |
|
Bacteroidetes |
|
|
Actinobacteria |
Eubacteria: |
Spirochaetes |
|
Archaea: |
|
|
Bacteroidetes |
Actinobacteria |
|
|
Crenarchaeota |
|
|
Fibrobacteres |
Bacteroidetes |
|
|
|
|
|
Firmicutes |
Cyanobacteria |
|
|
|
|
|
Proteobacteria |
Firmicutes |
|
|
|
|
|
|
Proteobacteria |
|
|
|
|
|
|
Spirochaetes |
|
|
|
|
|
|
Thermotogales |
|
|
|
|
|
|
Archaea: |
|
|
|
|
|
|
Crenarchaeota |
|
|
|
|
|
|
Euryarchaeota |
|
|
|
|
|
|
|
|
|
|
|

Rigden DJ. Iterative database searches demonstrate that glycoside hydrolase families 27, 31, 36, and 66 share a common evolutionary origin with family 13. FEBS Lett. 2002, 523(1-3):17 22.
GH-H
GH-D
кланы
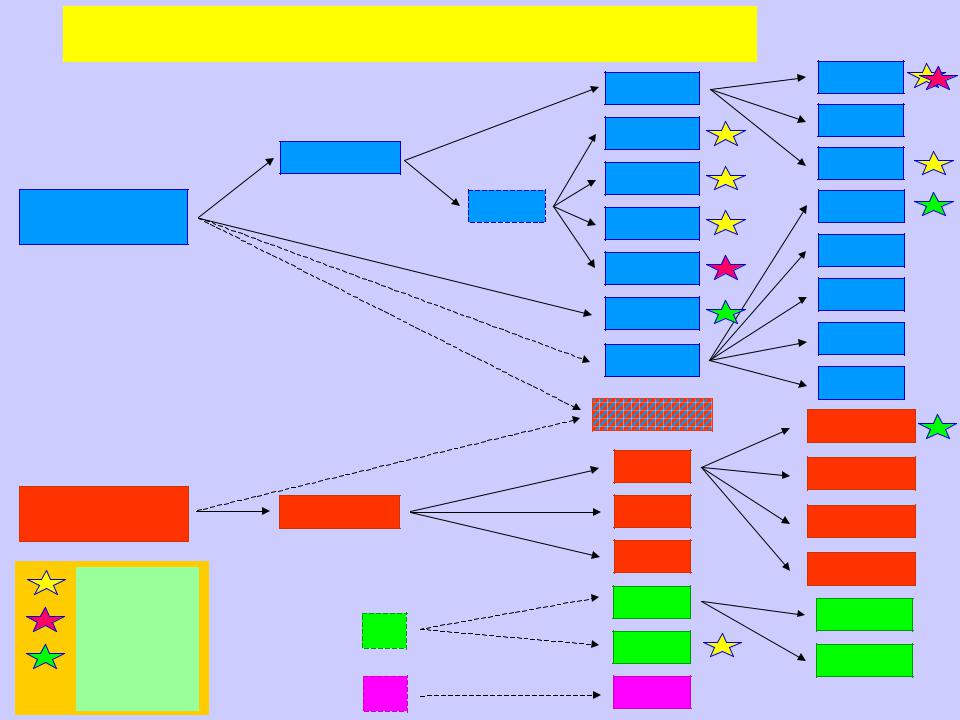
(β/α)8-barrel-type retaining α D-glycopyranosidase families
α galactosidase superfamily
α glucosidase superfamily
– [EC 3.2.1.22]
– [EC 3.2.1.49]
– [EC 3.2.1.20]
? – ORF
|
GH27 |
GH27a |
|
|
|
|
GH36A |
GH27b |
|
|
|
clan GH-D |
GH36B |
GH27c |
|
|
|
GH36 |
GH36C |
GH97a |
|
|
|
|
GH36D |
GH97b |
|
|
|
|
GH31 |
GH97c |
|
|
|
|
GH97 |
GH97d |
|
GH97e |
|
|
|
|
|
COG1649 ? |
COG0296 |
|
|
|
|
GH13 |
COG0366 |
|
|
|
clan GH-H |
GH70 |
COG1523 |
|
|
|
|
GH77 |
COG3280 |
|
|
|
|
GH38 |
family II |
? |
|
|
GH57 |
|
|
|
family III |
|
|
|
|
? |
GH66 |
|
?
?
?
?
?

Родственные связи (β/α)8 гликозидаз (Nagano et al., 2001)
Nagano N, Porter CT, Thornton JM. The (β/α)8 glycosidases: sequence and structure analyses suggest distant evolutionary relationships. Protein Eng. 2001, 14(11):845-855.
кланы: |
|
GH-H |
|
GH-A |
? |
GH-K |
|
|
|
|
|
|
|

Объединение семейств гликозидаз в кланы
Клан |
Семейства (GH) |
Оптическая конфигурация |
Трёхмерная структура |
|
|
|
|
GH-A |
1, 2, 5, 10, 17, 26, 30, 35, 39, |
сохраняется (экв.) |
(β/ )8 |
|
42, 50, 51, 53, 59, 72, 79, 86 |
|
|
GH-B |
7, 16 |
сохраняется (экв.) |
β-jelly roll |
|
|
|
|
GH-C |
11, 12 |
сохраняется (экв.) |
β-jelly roll |
|
|
|
|
GH-D |
27, 36 |
сохраняется (акс.) |
(β/ )8 |
|
|
|
|
GH-E |
33, 34, 83 |
сохраняется (экв.) |
6-fold β-propeller |
|
|
|
|
GH-F |
43, 62 |
меняется (экв.) |
5-fold β-propeller |
|
|
|
|
GH-G |
37, 63 |
меняется (акс.) |
неизвестна |
|
|
|
|
GH-H |
13, 70, 77 |
сохраняется (акс.) |
(β/ )8 |
|
|
|
|
GH-I |
24, 46, 80 |
меняется (экв.) |
+β |
|
|
|
|
GH-J |
32, 68 |
сохраняется (β фуранозид) |
5-fold β-propeller |
|
|
|
|
GH-K |
18, 20 |
сохраняется (экв.) |
(β/ )8 |
|
|
|
|
GH-L |
15, 65 |
меняется (акс.) |
( / )6 |
|
|
|
|
GH-M |
8, 48 |
меняется (экв.) |
( / )6 |
|
|
|
|
GH-N |
28, 49 |
меняется (акс.) |
(β)3-solenoid |
|
|
|
|

Объединение семейств гликозидаз в кланы
Клан |
Семейства (GH) |
Оптическая конфигурация |
Трёхмерная структура |
||
|
|
|
|
|
|
GH-A |
1, 2, 5, 10, 17, 26, 30, 35, 39, |
сохраняется (экв.) |
(β/ )8 |
|
|
|
42, 50, 51, 53, 59, 72, 79, 86 |
|
|
|
|
GH-B |
7, 16 |
сохраняется (экв.) |
|
β-jelly roll |
Strohmeier |
GH-C |
11, 12 |
сохраняется (экв.) |
|
β-jelly roll |
et al. (2004) |
|
|
||||
GH-D |
27, 36 |
сохраняется (акс.) |
(β/ )8 |
|
|
|
|
|
|
|
|
GH-E |
33, 34, 83 |
сохраняется (экв.) |
6-fold β-propeller |
|
|
|
|
|
|
|
|
GH-F |
43, 62 |
меняется (экв.) |
5-fold β-propeller |
|
|
|
|
|
|
|
|
GH-G |
37, 63 |
меняется (акс.) |
неизвестна |
|
|
|
|
|
|
|
|
GH-H |
13, 70, 77 |
сохраняется (акс.) |
(β/ )8 |
|
|
|
|
|
|
|
|
GH-I |
24, 46, 80 |
меняется (экв.) |
+β |
|
|
|
|
|
|
|
|
GH-J |
32, 68 |
сохраняется (β фуранозид) |
5-fold β-propeller |
|
|
|
|
|
|
|
|
GH-K |
18, 20 |
сохраняется (экв.) |
|
(β/ )8 |
|
|
|
||||
|
|
|
|
|
|
GH-L |
15, 65 |
меняется (акс.) |
|
( / )6 |
? |
|
|
||||
|
|
|
|
|
|
GH-M |
8, 48 |
меняется (экв.) |
|
( / )6 |
|
|
|
||||
|
|
|
|
|
|
GH-N |
28, 49 |
меняется (акс.) |
(β)3-solenoid |
|
|
|
|
|
|
|
|
