
ДНК-нанотехнологии 1 введение и основные методы / acs.chemrev.0c00294
.pdf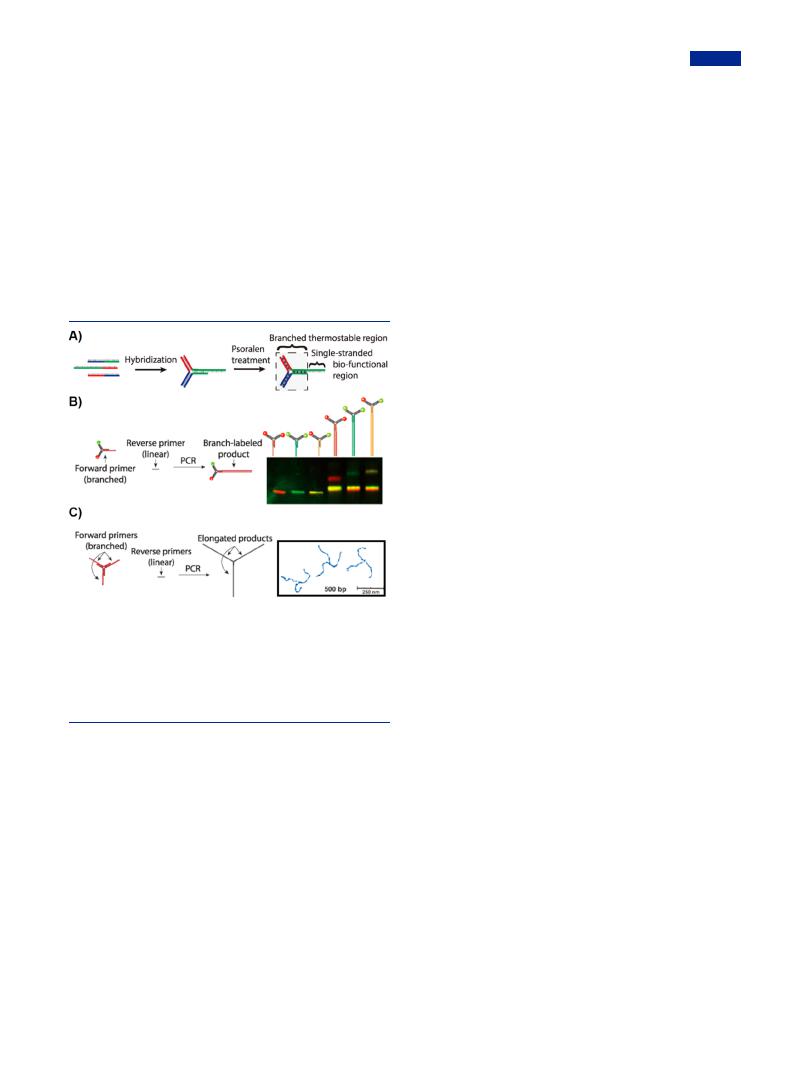
Chemical Reviews |
pubs.acs.org/CR |
Review |
PCR to prepare stable and condensed gene nanoparticles with multiple target gene cassettes together, which acted as transcription vectors for e cient protein expression and gene delivery in living cells.192 In 2016, Finke et al. used branched DNA that covalently linked with a cyclic peptide by click chemistry as primers of branched PCR to generate a new functional DNA-peptide network for specific cell attachment and culture.88
Psoralen was capable of stabilizing the branched DNA nanostructures. Upon UV irradiation, the compound could intercalate into the T bases and locked the double helical regions, thus enhancing the thermal stability of DNA constructions.193 Using this organic molecule, Hartman et al. developed thermostable branched DNA as modular primers for PCR, which remained intact under denaturing conditions such as high temperature and denaturing bu er (Figure 8).87 In
Figure 8. Enzymatic extension for the construction of branched DNA with ultralong arms. (A) Scheme of thermostable branched DNA as PCR primers. (B) Modular primers labeled with multiple fluorophores (left); gel electrophoresis demonstrating multiplexed PCR products with di erent combinations of FAM (green) and Cy5 (red) fluorophores. (C) Extension of Y-DNA by branched PCR (left); AFM images of branched elongated PCR products (right). Adapted with permission from ref 87. Copyright 2013, Wiley-VCH.
detail, the branched DNA contained a Y-shaped region and a single-stranded primer region. After treatment with psoralen, a high proportion of cross-linking sites formed in the doublestranded core region, which led the Y-shaped branch remain chemical stability at temperatures even higher than 95 °C. Meanwhile, the single stranded primer region remained its biological activity as a substrate for DNA polymerase (Figure 8A). By using the thermostable branched DNA as primers, branched PCR products were obtained in the process of PCR. The anisotropy branched DNA primers were modified with multiple fluorophores, resulting in a series of PCR products with specific colors (Figure 8B). Furthermore, when sticky ends of thermostable Y-shaped DNA were all designed as primer regions, elongated branched DNA products were obtained through PCR (Figure 8C). The chemically cross-linked branched DNA took advantages of both the thermostable assembly and intrinsic biological capabilities in a simple PCR process, and this technology created a versatile platform to
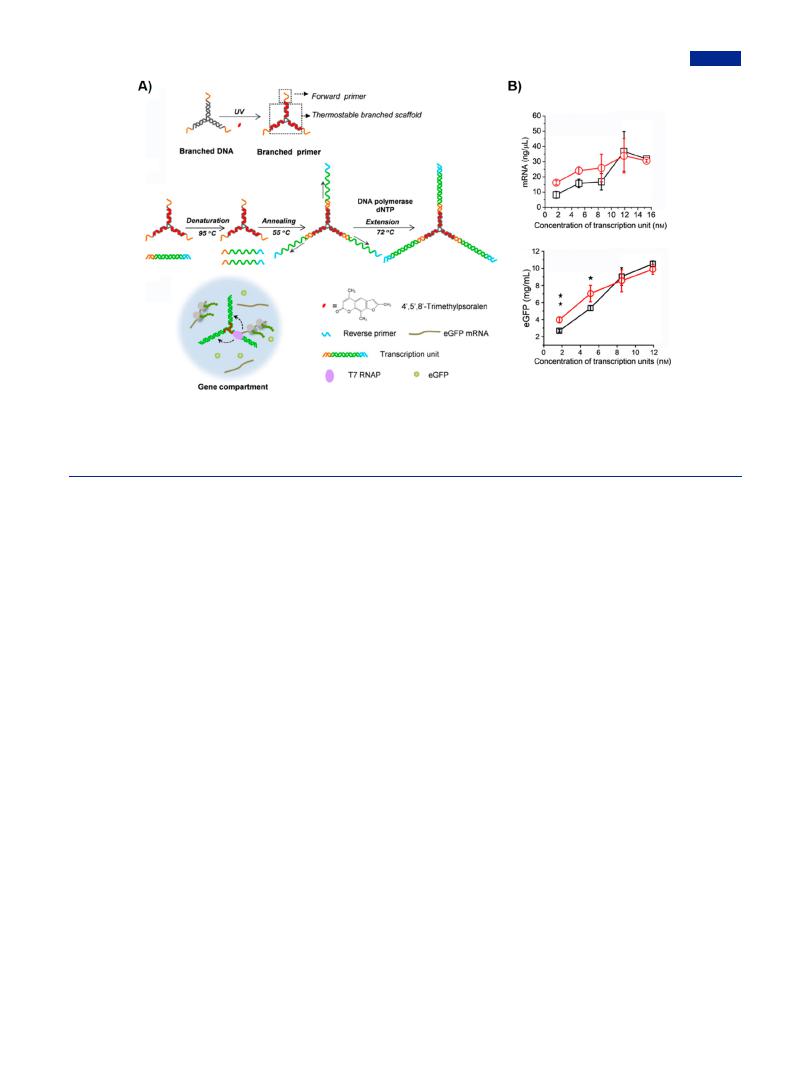
produce multifunctional DNA materials for a variety of applications.194 By using the branched PCR, Yang group constructed branched gene architectures as membrane-free gene compartment for studying proximity e ect in cell-free protein expression system (Figure 9).195 In single branched gene architecture, three transcription units were integrated by branched PCR, which enabled the RNA polymerase to shuttle more e ciently between neighbored units than free units (Figure 9A). The branched gene system increased local concentrations of DNA and RNA polymerase, resulting in improved gene expression yield with increasing concentration of transcription units and crowding agents (Figure 9B). The branched gene platform provided a strategy for studying the e ect of cellular crowed microenvironment on gene transcript and translation.
3.CONSTRUCTION OF ARTIFICIAL BRANCHED DNA-BASED FUNCTIONAL MATERIALS
Great progress in DNA nanotechnology has demonstrated that artificial branched DNA monomers as building-blocks exhibit the tremendous values in the fabrication of an assortment of functional DNA materials. According to the assembly principle, the methods for the construction of branched DNA-based materials were classified as tile-mediated assembly, DNA origami, dynamic assembly, and hybrid assembly.
3.1. Tile-Mediated Assembly
Tile-mediated assembly relied on branched DNA monomers as tile motifs that connected into high-level structures through the bottom-up hybridization of single-stranded overhangs. In tilemediated assembly, the structural and mechanical diversity of di erent branched DNA monomers were suitable for di erent types of structures. Branched DNA with multihelix as an arm, multipoint-star motif and DNA tensegrity triangle, had the characteristic of intermolecular tensegrity and fixed interbranch angle; consequently, they were more likely to be considered as tiles for the assembly of well-defined shape of wireframe structures. Wireframe structures containing hollow grid of geometric primitives were presented with the skillful combination of the flexible ssDNA as vertices and rigid dsDNA regions,196−198 including DNA arrays, polyhedrons and crystals. These highly ordered wireframe structures have shown merit as fundamental templates for the precise arrangement of nanoparticles and biomacromolecules due to predictable and addressable sites provided. Compared to the two branched DNA monomers, branched DNA with single-helix arm as tiles had relative flexibility and was amenable to the preparation of irregular DNA materials such as DNA dendrimers and DNA hydrogels.
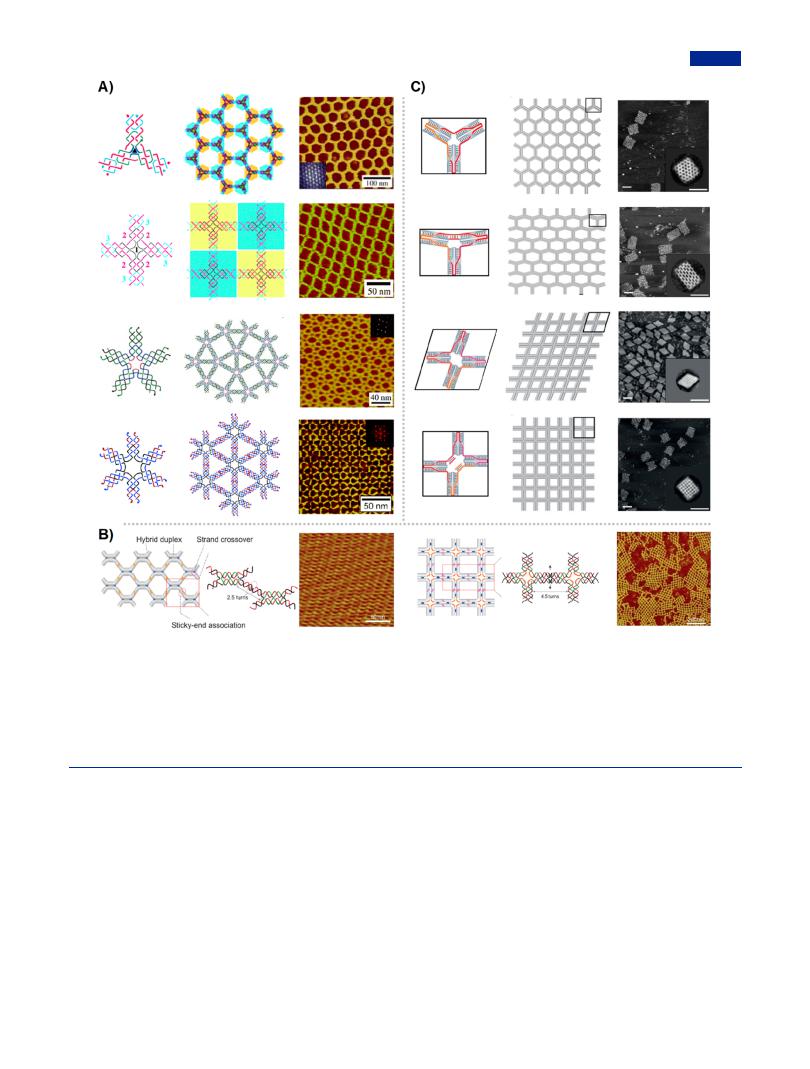
Modular assembly as a representation of tile-mediated assembly opened up new avenues to implement the design and construction of 2D DNA arrays from a family of multipointstar branched DNA.199,200 The periodic DNA arrays with di erent lattice patterns was attributed to the precisely designed branched DNA with diverse rotational symmetry, for example, 2D hexagonal, quadrangular, tetragonal, and triangular DNA arrays were constructed from three-point-star, four-point-star,
five-point-star, and six-point-star branched DNA, respectively (Figure 10A).58,201−203 The central loop was designed to
prevent the base stacking, and the sticky ends could be designed as palindromic sequences or symmetry−asymmetry pairs for end elongation and simplified operation;201 meanwhile, the interconnected junctions assembled from two branched DNA
9430 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 9. Branched PCR-based branched gene system for the construction of gene compartment. (A) Schematic of branched gene system constructed from thermostable branched primer and PCR-based assembly, which was employed as gene compartment to explore the e ect of gene proximity on gene transcription and protein expression in cell-free system. (B) Gene expression (including mRNA expression and protein expression) a ected by di erent concentrations of transcription, in which gene expression of branched genes showing up-regulation expression at a low concentration owing to gene compartmentation. Adapted with permission from ref 195. Copyright 2019, Wiley-VCH.
forced the DNA duplex to keep on a plane. In addition, to ensure the plane structures extended from multipoint-star branched DNA, a corrugation strategy was applied that neighboring units was linked by two-folded rotation axis (face-to-opposite sides of a plane) to eliminate the deviation of dihedral angle, determining the resulting plane structures.58 Note that the use of corrugation strategy was not amenable to all kinds of branched DNA to construct 2D arrays, which was in close correlation to the rotational symmetry. When rotational symmetry was 6-fold, which was not compatible with twofolded rotational symmetry, the neighboring units needed to face the same side to cancel existing curvatures.203 Interestingly, the subtle balance of conformational flexibility and stress, namely fine-tuning tensegrity, greatly influenced the stability, rigidity, and geometric structures of obtained assemblies, which were precisely adjusted by controlling the length of loop, sticky ends, and rigidity joints, as well as the duplex turnover number and interhelical angle.204−209 The bottom-up modular selfassembly was also amenable to the fabrication of DNA-RNA
hybrids branched motifs as programmable building-blocks and further 2D DNA arrays (Figure 10B).210 The fine and highly
ordered DNA arrays provided versatile platforms for the multiplexed and sensitive diagnostics of biomolecules, and information coding.211−213 Recently, a sca old-free “LEGO” approach to construct wireframe DNA arrays entirely from short strands was developed.214 In this approach, branched motifs were designed as two parallel duplexes with intersection at vertices instead of traditional intersection in the middle of duplexes, and three or four edges were diverged from a vertex. These subtle designs contributed to the formation of lattice-free wireframe DNA structures, including 2D arrays with honeycomb, rhombic, T-shaped, and cross-shaped patterns (Figure 10C). Furthermore, the flexible conformation could be achieved by adjusting the joint angels through the incorporation of ssDNA or dsDNA linkers. The sca old-free approach was also
employed to build fully addressable wireframe DNA architectures composed entirely of single-stranded tiles such as 2D arrays with hexagonal, chimeric, and cobweb-like tessellation patterns.72 These supramolecular 2D arrays could be covalently linked by photoactive anthracene dimerization to construct 2D polymers in solution, which exhibited solvent responsivity and structural transitions owing to the amphiphilic nature of organic molecules.185
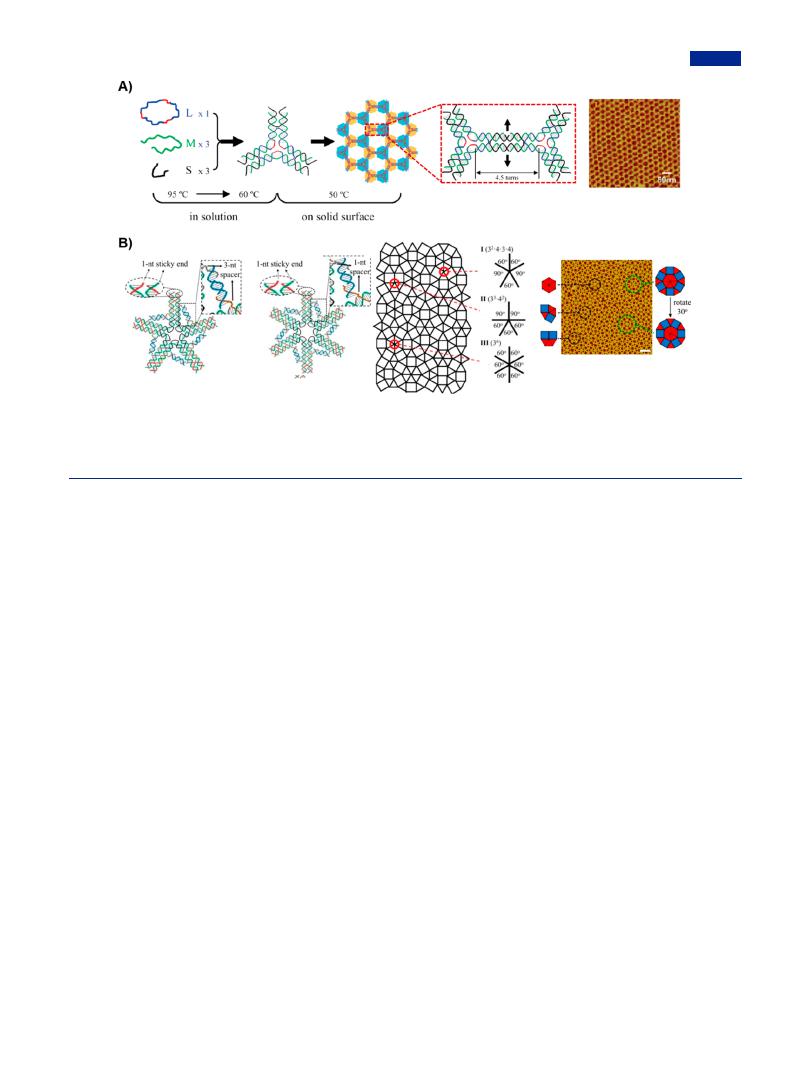
Additionally, surface-mediated self-assembly that was first proposed in 2009 was considered as an alternative to assembly in solution (Figure 11A).215 As a demonstration, the 2D DNA arrays were synthesized directly in situ on the surface of mica. During the self-assembly process, temperature as the essentially important parameter, needed to be controlled within the appropriate range, which required a comprehensive consideration of the balancing states of individual tiles and the entire array. The selection of appropriate temperature facilitated to the interaction of DNA tiles to mica surface and e ciently circumvent kinetic barrier.216 Compared with solution assembly, surface-mediated assembly showed advantages including eliminating the process from sample preparation to characterization and preventing the formation of 3D aggregations. It was noteworthy that surface-mediated assembly provided the opportunities in the building of irregular 2D arrays by the heterogeneous assembly of discrepant branched DNA motifs. These branched DNA motifs were designed with di erent finetuned interbranch angel and precisely regulated length of multibranches, and followed by depositing on solid surface to form irregular DNA arrays via blunt-end stacking (Figure 11B).
Numerous attempts have been made to demonstrate that tilemediated assembly has great potential in building custom 3D DNA polyhedrons from branched DNA, in particularly multipoint-star branched DNA.125,218 Because the entirety of a vertex and adjacent edges in DNA polyhedron could be regarded as branched DNA monomer, DNA polyhedron was the
9431 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 10. Tile-mediated assembly for the construction of 2D DNA arrays. (A) Multibranched DNA nanostructures with sticky ends as the building units for the construction of di erent classes of 2D DNA arrays, including hexagonal 2D DNA array from three-point-star DNA motif (Adapted with permission from ref 58. Copyright 2005, American Chemical Society), quadrangular 2D DNA array from four-point-star DNA motif (Adapted with permission from ref 201. Copyright 2005, Wiley-VCH), tetragonal 2D DNA array from five-point-star DNA motif (Adapted with permission from ref 202. Copyright 2008, National Academy of Sciences of the USA), and triangular 2D DNA arrays from six-point-star DNA motif (Adapted with permission from ref 203. Copyright 2006, American Chemical Society). (B) 2D DNA lattices reconstructed from DNA-RNA hybrid crossover tiles and DNA-RNA hybrid four-point-star DNA motif, respectively. Adapted with permission from ref 210. Copyright 2010, Springer Nature. (C) Addressable 2D DNA wireframe structures self-assembled from branched DNA motifs through precise angel control. Scale bars, 100 nm. Adapted with permission from ref 214. Copyright 2019, Wiley-VCH.
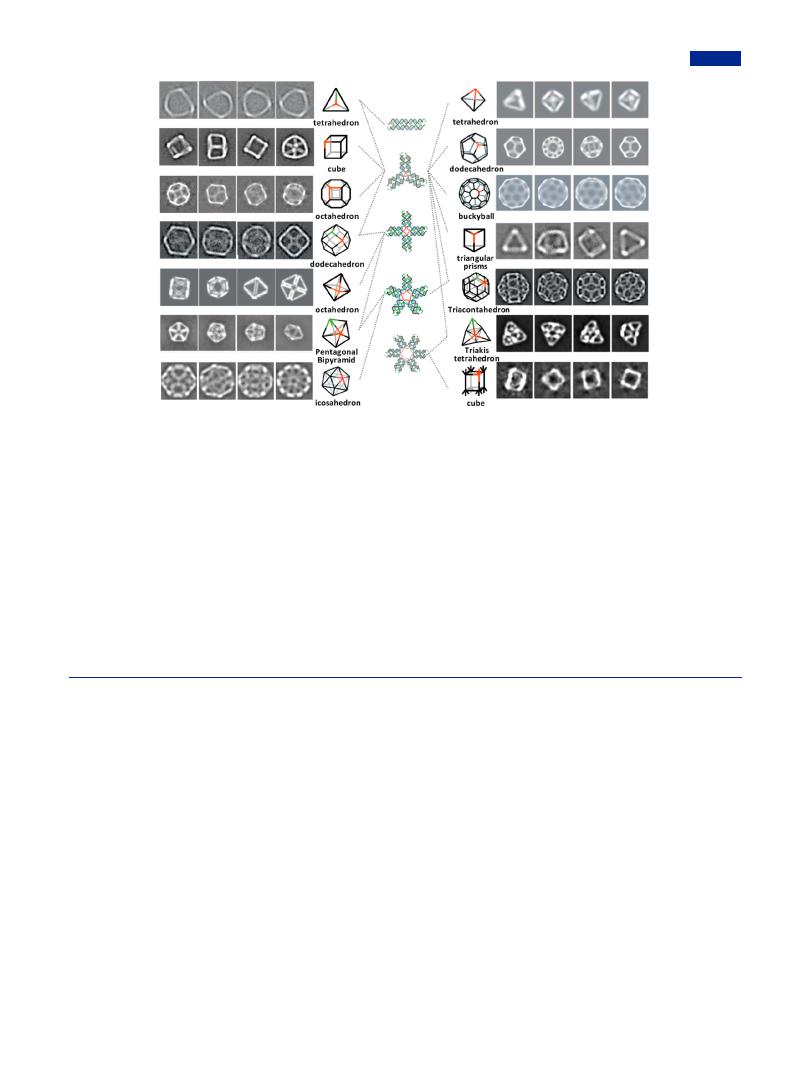
connectome of dendritic DNA by advantage of hierarchical assembly concept. The diversities of branched DNA in sequences, length, angles, and directions made them have a probability to assemble polyhedrons with multiple combinations (Figure 12) including same polyhedron from di erent types of motifs, individual polyhedron from copies of identical branched DNA, and single polyhedron from coassembly of multiple branched DNA.219,220 Typically, the branched points of branched DNA served as vertexes interconnected by sticky ends on edges, resulting in well-defined DNA polyhedron. The length and curvatures of DNA duplex needed to be adjusted to satisfy the requirement that all component branched DNA faced toward the same direction to form polyhedrons instead of planar structures. Meanwhile, lower branched DNA concentrations and more flexibility of loops by increasing bases number were
both prone to the formation of polyhedron.60 Assembled morphologies of DNA polyhedrons were tightly in association with the concentrations, flexibilities, and rigidities of the corresponding branched DNA monomers.202,221 By slightly changing the length of central loop, the relative inclination between the center and tile plane could be controlled, eventually forming the isomeric DNA polyhedron with inward or outward bending.222 The formation of chiral DNA structures also could be achieved by removing the rotation symmetry of DNA motifs on purpose.126 In addition, exterior modifications, such as hairpin, could be decorated at the addressable sites on DNA polyhedron for easily introducing appendage groups (i.e., component proteins).223 As an alternative of traditionally used sticky ends, T-junctions could also play a role in connection with neighborhood branched DNA, facilitating the formation of
9432 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 11. Surface-mediated self-assembly for the construction of 2D DNA arrays. (A) Surface-mediated assembly consisted of two steps: the formation of three-point-star motifs as tiles by solution annealing and 2D nanoarrays on solid surface. Right was the AFM image of hexagonal 2D array. Adapted with permission from ref 215. Copyright 2009, American Chemical Society. (B) Dodecagonal DNA 2D quasicrystals constructed from two kinds of DNA motifs with di erent branch vertices by surface-mediated assembly, which was characterized by AFM on the right. Adapted with permission from ref 217. Copyright 2019, American Chemical Society.
DNA polyhedrons with hairpins or aptamers for wider applications.224
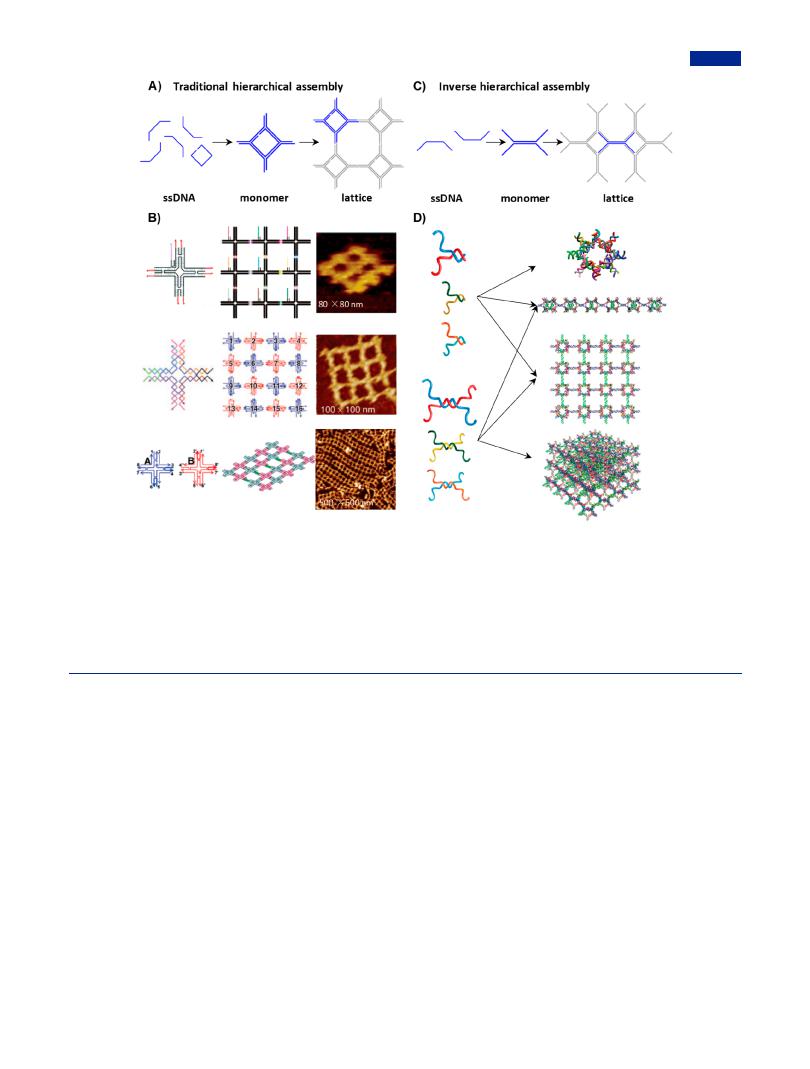
Additionally, a strategy of hierarchical assembly was exploited to construct fully addressable DNA arrays. In hierarchical subtile strategy, branched DNA tiles were first self-assembled from tweezer-like subtiles with flexible loop in two steps, and then assembled into a family of programmable superstructures in a more facile and controllable step, which provided steric crowding and reduced the undesired pairing in one-pot method.226 In addition, a stepwise hierarchical assembly was developed to construct DNA arrays with finite boundary (Figure 13A), which addressed the issue that spatial positioning was inaccurate in DNA array without preset boundaries. The stepwise hierarchical assembly began with the branched DNA tiles as monomers linked by sticky ends, and preformed smallsized lattices and subsequent enlarged arrays in sequential steps. The obtained DNA arrays as molecular pegboard allowed for the specific locations of biomolecules with nanoscale accuracy (Figure 13B).229 There were two extreme cases in hierarchical self-assembly: the minimum number of assembly units and the minimum number of assembly steps (also called depth). Taking a 4 (row) × 4 (column) DNA lattice as an example, eight sequence sets each consisting of two motifs, and four sequential steps were required in the case of minimum number of assembly units. When the case of minimum depth was applied, a sequence set containing 16 motifs with unique sticky ends and one-step equimolar mixing were needed. The hierarchical assembly technique reliably controlled each step of assembly at moleculelevel, so that arbitrary finite-size DNA arrays were guaranteed to obtained.226 The production yield was increased by the compromise between two extreme cases. In addition, the incorporation of dsDNA nanobridges joining adjacent tiles was used for regulating the length and directionality of DNA arrays (Figure 13B).230 The fully addressable and finite-size DNA
arrays allowed the accurate positioning of specific biomolecules in nanosized space.229
Contrary to the assembly order of traditional hierarchical assembly, inverse hierarchical assembly performed with joint wires as beginning modules to form branched junctions (Figure 13C).231 The number of composition chains involved in inverse hierarchical assembly was significantly less than that in hierarchical assembly; moreover, the strategy of inverse hierarchical assembly was conducive to the formation of ultrabranched junction with centrosymmetric topology. In addition, to prove the universality of the inverse strategy in constructing complex DNA nanostructures, two kinds of connection wires were in the attempt designed at first, including half double-stranded connection wires and double-stranded connection wires. Following the inverse hierarchical assembly, stable zero dimensional (0D), one-dimensional (1D), 2D, and 3D DNA arrays could be assembled by the ingenious combination of two kinds of connection wires (Figure 13D).
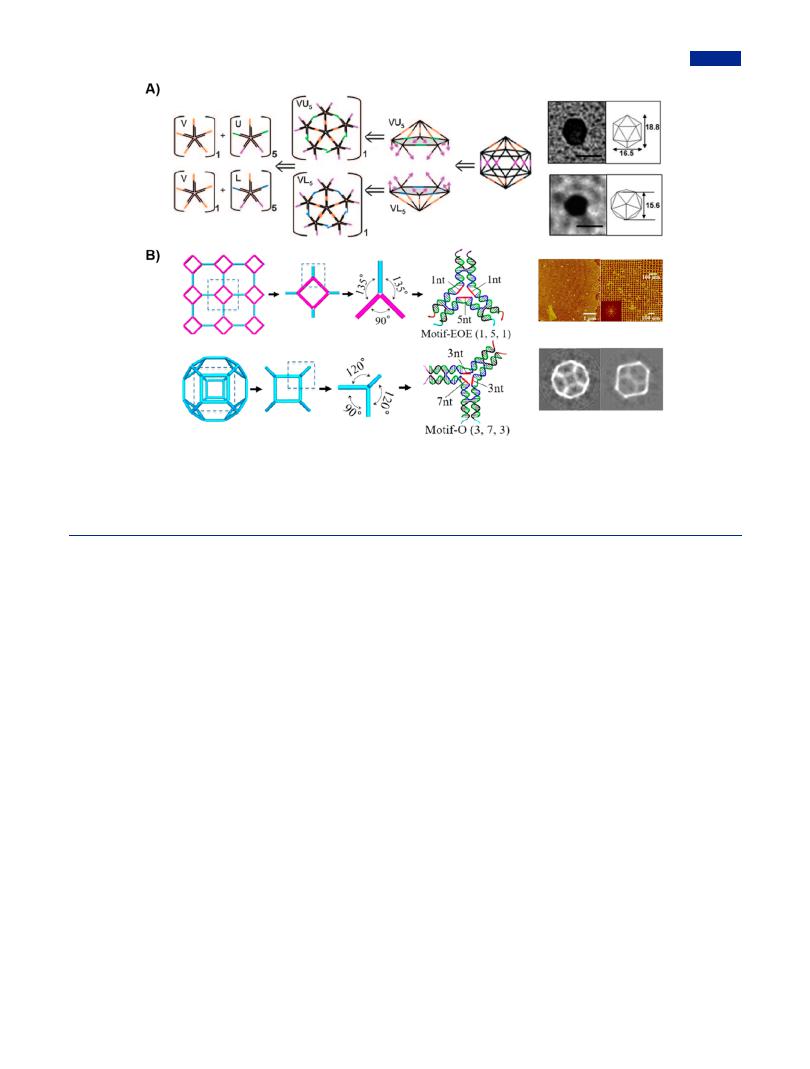
Inspired by converse thinking, retrosynthetic strategy was first reported by Krishnan group for the nanofabrication of DNA icosahedrons.63 Retrosynthetic strategy was by definition to start with the desired assembly, which was divided into basic modules in the top-down principle. These modules as building units could assemble into expected nanostructures in a predesigned manner. The construction strategy minimized the trial and error process, greatly shortening the experimental period. Taking DNA icosahedron as an example, it was split into two halficosahedra on the basis of retrosynthetic strategy, and subsequently the half-icosahedra was divided into two kinds of five-armed branched DNA in a molar ratio of 1:5. The programmable overhangs on two half-icosahedra were complementary to each other to fabricate an integrated icosahedron (Figure 14A). The obtained DNA icosahedron had a large volume of cavity and a dense surface, which thus had great potential as a “smart” nanocapsule in the encapsulation and
9433 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 12. Hierarchical assembly for the construction of DNA polyhedrons. Various DNA polyhedrons could be assembly by single category of branched DNA, such as buckyball from three-armed branched DNA, icosahedron from five-armed branched DNA and cube (seventh from right) from six-armed branched DNA. Some identical polyhedrons could be synthesized by di erent branched DNA monomers; for instance, octahedron (third and fifth from left) could be prepared by three-armed branched DNA and four-armed branched DNA, respectively. In addition, some DNA polyhedrons were required to be constructed with participation of multiple types of branched DNA, such as pentagonal bipyramid from four-armed and five-armed branched DNA chimeras, triacontahedron from three-armed and five-armed branched DNA chimeras, and triakis tetrahedron from three-armed and six-armed branched DNA chimeras. Some of these DNA polyhedrons could be assembled by either single category of branched DNA or a combination of multiple categories of branched DNA. For example, one tetrahedron that was the first from right could be individually constructed from three-point-star branched DNA, and another one that was the first from left could also be constructed from the combination of three-armed branched DNA and dsDNA. Similarly, dodecahedron (one was the second from right and another was the fourth from left) was respectively composed of three-armed branched DNA, and three-armed and four-armed branched DNA chimeras. The reconstructed 2D projections of tetrahedron (first from left), dodecahedron (fourth from left) and triacontahedron were reprinted with permission from ref 219. Copyright 2017, Elsevier Publishing Group; of cube (second from left) was reprinted with permission from ref 221. Copyright 2009, American Chemical Society; of octahedron (third from left) was reprinted with permission from ref 128. Copyright 2016, American Chemical Society; of octahedron (fifth from left) was reprinted with permission from ref 222. Copyright 2010, Wiley-VCH; of pentagonal bipyramid was reprinted with permission from ref 220. Copyright 2014, WileyVCH; of icosahedron was reprinted with permission from ref 202. Copyright 2008, The National Academy of Sciences of the USA; of tetrahedron (first from right), dodecahedron (second from right) and buckyball were reprinted with permission from ref 60. Copyright 2008, Springer Nature; of triangular prisms was reprinted with permission from ref 126. Copyright 2012, Wiley-VCH; of triakis tetrahedron and cube (seventh from right) were reprinted with permission from ref 225. Copyright 2015, Wiley-VCH.
controlled release of cargoes, especially biomacromolecules possessing relatively large molecular size.232 For example, a kind of fluorescent biopolymer as model cargoes was encapsulated into the nanocapsules during nanofabrication. Moreover, responsive modules enabled the controlled opening of nanocapsules in the presence of corresponding external triggers, finally generating fluorescence signals with enhanced intensity.233 This platform based on icosahedral DNA nanocapsules was available to study the release kinetics of signal conducting molecules under chemical trigger. In addition, by combining with other functional components, such as target modules, quantum dots, and endocytic ligands, multifunctionalized DNA nanocapsules enabled to accomplish the targeted delivery, stimuli-triggered cargo release with spatiotemporal precision, spatial pH mapping of endosomal maturation, and real-time tracking of endocytic pathways.234−237 These researches showed enormous values of “smart” DNA icosahedra-based nanocapsules in the territory of biomedicine.
Another advantage of retrosynthetic strategy was the greater tolerance for the symmetry of branched DNA, resulting in a more flexible configuration of branched DNA. The proposal of
retrosynthetic strategy broke intrinsic constraint of using highly symmetric branched DNA in conventionally used approaches. Wang et al. made use of the strategy to guide the assembly of complex DNA polyhedrons (Figure 14B).128 Three types of DNA motifs with di erent combinations of helical number were designed including branches all with odd-numbered helixes, branches all with even-numbered helixes, and chimeric branches with oddor even-numbered helixes. Accordingly, four rules for the assembly based on asymmetry tiles were proposed: (1) oddnumbered helixes as edges intended to pattern corrugated assemblies instead of closed rings, while edges containing evennumbered helixes promoted cyclization; (2) central loops extension contributed to the out-of-plane flexibility of DNA motifs with high degree; (3) the loop length showed a reverse correlation with interbranch angle; that is, the long loop favored a small angle between flanking branches; (4) higher concentrations of DNA motifs preferred to larger size of DNA structures. These fundamental rules were expected to be promising guidelines for the synthesis of complex DNA polyhedrons from various branched DNA.
9434 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 13. Traditional hierarchical assembly and inverse hierarchical assembly. (A) Scheme of traditional hierarchical assembly, where branched junctions served as monomers first and then self-assembled into 2D nanoarrays by the connection of sticky ends.231 (B) Finite-size 2D DNA nanoarrays formed by stepwise hierarchical assembly, including molecular pegboard (Adapted with permission from ref 229. Copyright 2005, American Chemical Society), 4 × 4 lattice from 16 DNA cross-tiles (Adapted with permission from ref 227. Copyright 2005, Wiley-VCH) and heterogeneous DNA lattices with dsDNA bridges from two types of DNA tiles (Adapted with permission from ref 230. Copyright 2008, American Chemical Society). (C) Scheme of inverse hierarchical assembly, in which connection wires were formed as first, and then self-assembled into branched junctions in lattice. (D) Diverse combinations of half double-stranded connection wires and double-stranded connection wires as tiles for the assembly of 0D, 1D, 2D and 3D lattices. For example, 0D and 3D lattices were totally formed by half double-stranded connection wires and doublestranded connection wires, respectively. 1D and 2D lattices were composed of the di erent combinations of two kinds of connection wires. Adapted with permission from ref 231. Copyright 2017, American Chemical Society.
Under the guidance of tile-mediated assembly, the rigid and well-structured branched DNA as building units could construct macroscopic 3D DNA crystals, breaking through the limitation of the nanoscale. Zheng et al. constructed macroscopic DNA crystals from DNA tensegrity triangle as building units.62 The tensegrity triangle consisted of three helical domains with three separated branch points, resulting in sti and periodic 3D crystals when extending from six directions (Figure 15A). Both the constitution of the building units and the interunit interactions were adjustable: the units could be functionalized by a triplex-forming oligonucleotide on each of its helical edges;111 the interunit interactions within the 3D DNA crystals could be enhanced by introducing a triplex-forming oligonucleotide at the interunit cohesion region.129 The weak lattice interactions and high concentrations were decisive factors for achieving the crystallization, while sample heterogeneity and impurities played a minor role.238 Besides, the crystallization process responded to temperature, which allowed the control of the system.239 The self-assembled crystal could in turn help to determine the structure of the unit such as incorporated stressed DNA within the crystal lattice.240 Wang et al. demonstrated the feasibility of using sticky-end directed self-assembly to control
the contents of a macromolecular 3D crystal.241 Two di erent triangle molecules were designed, and the only intermolecular interactions between the molecules were programmed with sticky ends; as the asymmetric units, the triangle molecules alternated through the lattice within the crystal (Figure 15B). In addition, the covalent attachment of dye molecules to the DNA units resulted in di erent colors of the crystals. Energy transfer in the network of dye-attached DNA crystal could be studied.242 Great attempt have been made to control the crystallization kinetics and improve the quality of engineered 3D DNA crystals. The e ect of varying sticky end lengths and sequences as well as the impact of 5′- and 3′-phosphates on the improvement of crystal formation and resolution were investigated.243,244 Another e ective strategy was introducing a rationally designed agent to modulate the crystallization process, by which means, larger crystals with higher resolution were produced.245 To overcome the constraint of stability of DNA crystals, an approach to prepare robust 3D DNA crystals by postassembly ligation was recently developed (Figure 15C).246 Sticky ends of DNA fragments at crystal contacts were enzymatically ligated, and the covalent bonds significantly enhanced crystal stability (Figure 15D).
9435 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 14. Retrosynthetic strategy for the construction of complex DNA nanostructures. (A) DNA icosahedron was constructed from two kinds of half-icosahedron, VU5 and VL5, which were self-assembled by five-armed branched DNA with complementary overhangs. The transmission electron micrographs of DNA icosahedron were shown on the right. Adapted with permission from ref 63. Copyright 2009, Wiley-VCH. (B) Retrosynthetic analysis for the assembly of complex DNA nanostructures from well-defined DNA motifs. The intermolecular branches of DNA motifs were rationally predesigned by the retrosynthetic analysis. On the right were the AFM images of square 2D lattice and the reconstructed projections of DNA octahedron. Adapted with permission from ref 128. Copyright 2009, Wiley-VCH.
More and more e orts have been made to create new types of branched DNA for constructing DNA crystals in tile-mediated assembly. The layered-crossover motif, which has been previously used to construct 3D framework DNA origami structures, was implemented to construct a set of rhombus-like layered-crossover DNA tiles with precisely tunable orientation angles, which further assembled into 3D crystals with dimensions up to hundreds of micrometers (Figure 15E).247 Simmons et al. demonstrated the structure of a new crystal system inspired by DNA tensegrity triangle. The motif consisted of three component strands self-assembled into designed crystalline arrays for each form, with a layered lattice mediated via a central strand that contained a sequence of 4 sequential repeats of 6 bases (Figure 15F).248 The left-handed enantiomer of this motif was crystallized to obtain two self-assembling DNA crystal systems. They further designed and crystallized the first example of a 3D DNA array with a layered hexagonal lattice (Figure 15G).249 Their approach provided a new avenue to create self-assembled macromolecular 3D crystalline lattices with atomic precision, which contained a highly organized array of well-defined cavities.
By adapting multistage hierarchical assembly, branched DNA with single-helix as an arm was used to construct DNA dendrimers. Generally, the structure of DNA dendrimers could be divided into three regions: the branched core, the repetitive units, and the end groups.162,250 Branched DNA monomers could act not only as the branched core but also as repetitive units to generate multiple layered DNA dendrimers.251,252 In general, the resulting DNA dendrimers had high synthetic productivity and excellent monodispersity, whose controllable sizes were regulated by monomers types and
concentrations, facilitating to size-dependent endocytosis. Relative to branched DNA monomers, DNA dendrimers possessed numerous prominent advantages such as larger surface area, more binding sites, and improved robustness in cellular microenvironment. In addition, the multiple end groups endowed DNA dendrimers with inherent properties of multivalency, modification, and functionality.253,254 These incomparable advantages made it widely applied in biomedical fields including multiplexed detection of biomolecules, drug delivery, and therapeutics.255
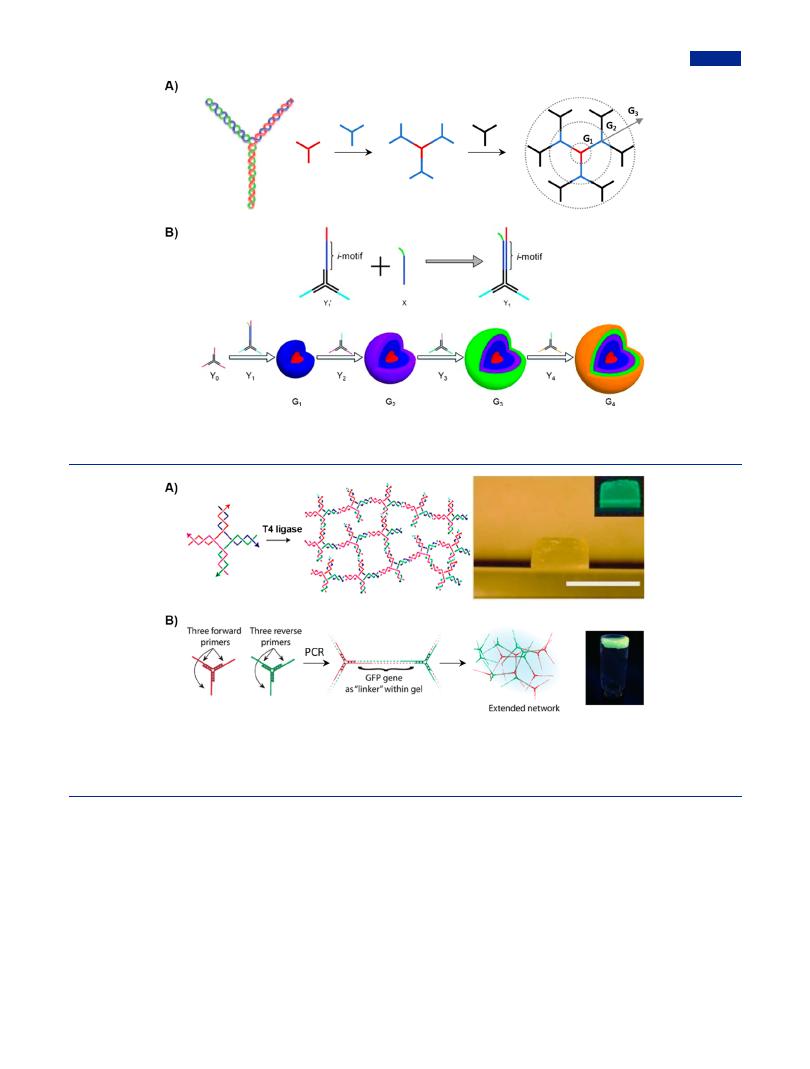
Relying on the complementary pairing, Li et al. achieved the controlled assembly of multigeneration DL-DNA structures with Y-shaped DNA as the branched core and repetitive units (Figure 16A).57 The sticky ends on Y-DNA were specially designed as nonpalindromic sequences to avoid self-ligation. And the design of sticky ends of Y-DNA followed the principles of “intra-layer consistency, inter-layer di erence”, to ensure the ordered assembly of DNA dendrimers in multistage hierarchical assembly. The resulting DL-DNA was obtained through repeatedly ligating peripheral Y-DNA to the sticky ends of the previous generation. The resulting dendrimer DNA showed stable and monodisperse structures, and di erent di usion behavior relative to linear DNA structures.94 The multiplex and anisotropic DL-DNA were developed as a fluorescent nano- barcodes-based platform for the multiplex detection such as pathogens DNA, ribonucleic acid (RNA), and protein.251 The size of DL-DNA structures was tunable not only by controlling the number of generations but also by introducing adjustable modules in response to corresponding stimulus. In addition, the introduction of pH-responsive modules (i.e., i-motif) into DNA dendrimers in a stepwise assembly caused DNA dendrimers to
9436 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 15. Tile-mediated assembly for the construction of DNA crystals. (A) Schematic of the branched DNA with three branched points (also called tensegrity triangle) as the building units of macroscopic 3D DNA crystals, and the optical image of DNA crystals showing rhombohedral shape. Adapted with permission from ref 62. Copyright 2009, Springer Nature. (B) Single crystal with controlled color formed by two di erent tensegrity triangles by adding Cy3 and Cy5 dyes. Adapted with permission from ref 241. Copyright 2010, American Chemical Society. (C) Robust DNA crystal by covalent bonding in replace of sticky-ended cohesion, overcoming a key obstacle of highly constrained conditions. (D) Covalent bonds-linked DNA crystal showing significantly enhanced robustness even in harsh environments, including 65 °C for 16 h, 42 days in water, as well as rehydrate after drying and butanol. Adapted with permission from ref 246. Copyright 2019, American Chemical Society. (E) Assembled 3D crystals from layeredcrossover branched DNA with di erent cross angle, 90° and 60°, respectively. Adapted with permission from ref 247. Copyright 2018, American Chemical Society. (F) Schematic (upper) and light microscope images (bottom) of four-layered D-DNA crystal and L-DNA crystal, which represented in orange and blue, respectively. DNA with right-handed helical twist was called as D-DNA, and its enantiomer was called as L-DNA. Adapted with permission from ref 248. Copyright 2017, American Chemical Society. (G) Schematic of 3D crystal from branched DNA between neighboring layers, which exhibited 6-fold symmetry at 90° rotation. Adapted with permission from ref 249. Copyright 2018, Wiley-VCH.
possess the characteristics of stimulus responsiveness and sizeadjustability (Figure 16B).97 Under weak acidic environment (pH 5.0), the i-motif conformation changed from linear to fourstranded structures, leading to a reduction in size. In particular, DNA dendrimers could e ectively protect single-stranded nucleic acid from degradation, which usually occurs in the cellular or plasma environments. Therefore, DNA dendrimers could be used as highly e cient and intracellular delivery nanocarriers of single-stranded nucleic acid.
Tile-mediated assembly has drawn great attentions as an outstanding strategy not only in the construction of nanoor microscale materials but also at macroscale, typically DNA hydrogels. The multidirection extension of DNA strands from branched DNA was used for the formation of cross-linked network in hydrogels. Meanwhile, the tunable rigidity and flexibly of branched DNA dominated the mechanical strength of DNA hydrogel, accompanied with shape plasticity or shape adaptability. DNA hydrogels have exhibited unique advantages such as mimicking extracellular matrix (ECM) environment and possessing appropriate mechanical strength; DNA hydrogels
therefore had prospective biological applications including sca olds for tissue engineering, wound dressing, 3D cell culture, capture and delivery, and protein production.256−263 Implementing enzymatic reactions was a general approach to synthesize branched DNA-based hydrogels,264 and enzymefree self-assembly was also adopted.265
The pure DNA hydrogel was first demonstrated by Um et al. based on branched DNA via enzymatic reactions (Figure 17A).59 Branched DNA termed as X-, Y-, and T-shaped branched DNA (namely X-, Y-, and T-DNA, respectively) hybridized and ligated with each other through covalent linkages among palindromic sticky ends, serving as both monomers and cross-linkers. In addition, the mechanical property of DNA hydrogels was controlled by the initial concentration and types of branched DNA monomers. Remarkably, the swollen DNA hydrogel constructed from X-DNA monomers showed better resistance to degradation and long-term drug release. The swollen degree and phase behaviors of DNA hydrogel were influenced by binding energy or hybridization strength, including the unpaired bases located in the sticky ends of X-
9437 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 16. Multistage hierarchical assembly for the construction of DNA dendrimers. (A) Schematic drawing of Y-DNA (upper) and higher generation DL-DNA (below). Adapted with permission from ref 57. Copyright 2004, Springer Nature. (B) DNA dendrimers with i-motif structures possessing the property of size-adjustability. Adapted with permission from ref 97. Copyright 2012, Wiley-VCH.
Figure 17. Tile-mediated assembly for the construction of pure branched DNA-based hydrogels via enzymatic reactions. (A) Schematic illustration of enzyme-catalyzed assembly of DNA hydrogel from branched DNA building-blocks (left); a swollen X-DNA hydrogel synthesized in a cylindrical mold (right, scale bar, 1 cm). The inset shows the DNA gel stained with SYBR Green I (right). Adapted with permission from ref 59. Copyright 2006, Springer Nature. (B) Formation of DNA hydrogel by branched PCR (left); photograph of DNA hydrogel stained with GelGreen under UV illumination (right). Adapted with permission from ref 87. Copyright 2013, Wiley-VCH.
DNA monomers, salt ion types, and temperature.266 Production of sticky ends could be achieved by enzymatic treatment, mainly by terminal deoxynucleotidyl transferase (TdT), which was capable to extend the poly(A) or poly(T) tails.267 Taking advantage of the hybridization of poly(A) and poly(T), branched DNA were interconnected into DNA hydrogel with high mechanical strength. Branched DNA integrated with forward and reverse primers was utilized to create ultralong chains in networks and further hydrogels implemented by the amplification of branched PCR, which encapsulated fluorescence protein gene and was useful in some biotechnological applications such as protein production (Figure 17B).87 By encapsulating lipid on the surface of enzyme-linked DNA
network with gene-knitted X-DNA as a monomer, a shell−core structure was prepared, which was speculated to be useful for in vitro protein production.268 By introducing conductive carbon nanotubes (CNTs) and polyaniline coating into the enzymelinked DNA hydrogel template, the functional DNA hybrid hydrogel was constructed as a supercapacitor with ultralow cytotoxicity.269
In addition to employing enzyme-assisted ligation to connect branched DNA together, enzyme-free self-assembly was also used to construct branched DNA-based hydrogels. As a tile, branched DNA was networked merely by DNA linkers to form hydrogels in tile-mediated assembly. On the one hand, enzymefree and tile-mediated assembly relying on hydrogen bonds
9438 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
Chemical Reviews |
pubs.acs.org/CR |
Review |
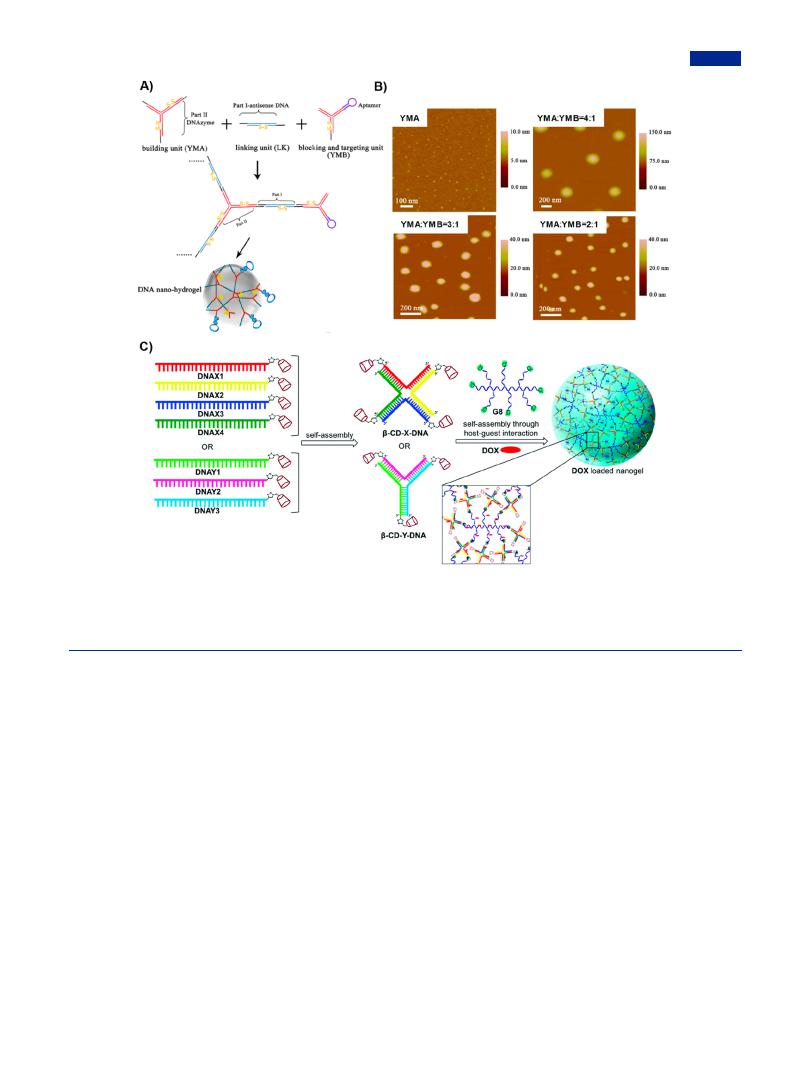
Figure 18. Tile-mediated assembly for the construction of branched DNA nanohydrogels. (A) DNA nanohydrogel self-assembled from extensible Y- shaped DNA (YMA), partially terminated Y-shaped DNA (YMB), and linking unit. (B) Size of DNA nanohydrogels regulated by adjusting the ratio of YMA and YMB. Adapted with permission from ref 66. Copyright 2015, American Chemical Society. (C) DOX-loaded DNA nanohydrogel selfassembled via multivalent host−guest interactions between β-CD modified branched DNA and adamantly grafted polymer. Adapted with permission from ref 278. Copyright 2017, Royal Society of Chemistry.
showed irreplaceable advantages in reversible dynamic assembly, evading the extremely stable performance of covalent bonds in enzyme-assisted ligation. This enzyme-free hydrogel was more conducive to regulating its thermodynamic properties.122,270 Meanwhile, the design of branched DNA as pieces of mosaic tiles facilitated the overall dissociation of hydrogels and the incorporation of functional elements. Some responsive elements embedded into sticky ends of branched DNA endowed the adjustable mechanical strength and repetitive gel-to-sol transition of hydrogels under the action of corresponding stimulus. For example, the hybridization of aptamer and complementary sequences induced the disintegration of DNA hydrogel with aptamer functionalized.271,272 A photoresponsive base had the capability of controlled disassembly of hydrogel under UV irradiation.273 The characteristic of this dynamic controlled disassembly, coupled with excellent biocompatibility, made DNA hydrogel suitable for in situ encapsulation and controlled release of proteins, while maintaining their activities.274,275 On the other hand, the enzyme-free process was advantageous to the adjustment of size of hydrogel, from macroscale to nanoscale, in a more controlled fashion, such as adding blocking chains. Regulating the concentration ration of branched DNA tiles, DNA linkers, and blocking chains
e ectively controlled the size of nanohydrogel. The nanosized DNA hydrogels occupied a dominant position in biological applications, and have drawn ever-increasing attention of researchers to develop new preparation strategies. In 2015, Tan and co-workers prepared the size-controllable and stimuliresponsive nanohydrogels based on the DNA self-assembly of Y- shaped DNA as monomers and dsDNA as linkers (Figure 18A).66 One Y-shaped monomer contained three sticky ends, while the other Y-shaped monomer contained a sticky end for extension, a blunt end and an aptamer for blocking. The size of DNA nanohydrogels was controlled by varying the ratio of two kinds of Y-shaped monomers (Figure 18B). The incorporation of other functional components, such as aptamers, therapeutic agents, and disulfide linkages, endowed the nanohydrogel with multiple performances, including the specific targeting mediated by aptamers and stimuli-responsive degradation to reductive microenvironment (i.e., endogenous glutathione) that was provided by disulfide linkages. In addition, by conjugating therapeutic antisense DNA and DNAzyme oligonucleotides, the DNA nanohydrogels showed e ective inhibition of cell proliferation and migration. Some nucleoside analogues, such as floxuridine (F) similar to thymidine (T) and gemcitabine (Ge) similar to deoxycytidine (dC), could be easily integrated
9439 |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. 2020, 120, 9420−9481 |
