
- •Лекция 1 Лекция 1
- •Edward Jenner "the father of immunology"
- •smallpox vaccine, the world's first vaccine.
- •Louis Pasteur
- •ОпределениеОпределениеиммунологиииммунологии
- •Главная функция иммунной системы – распознавание «свой-чужой»
- •Эволюция иммунных механизмов
- •ОсновныеОсновныеэлементыэлементыиммуннойиммуннойсистемсистем
- •ПроисхождениеПроисхождениеклетокклетокиммуннойиммуннойсистесист
- •ПроисхождениеПроисхождениеклетокклетокиммуннойиммуннойсистесист
- •сновныеновные лимфоидныелимфоидныеорганыорганыииобразованобразован
- •РециркуляцияРециркуляциялимфоцитовлимфоцитовии антигенантиген--презентирующихпрезентирующихклетокклеток
- •Поверхностные маркеры
- •ЕстественныеЕстественныеии адаптивныеадаптивные иммунныеиммунныемеханизмымеханизмы
- •ФагоцитарныеФагоцитарныеклеткиклетки..СистемаСистема мононуклеарныхмононуклеарныхфагоцитовфагоцитов
- •Функции комплемента в антибактериальном иммунитете
- •ФагоцитозФагоцитоз
- •ФагоцитозФагоцитоз
- •ВзаимодействиеВзаимодействиемеждумеждулимфоцитамилимфоцитами иифагоцитамифагоцитами
- •Клеточный иммунный ответ
- •Суперсемейство иммуноглобулинов – основа системы распознования «свой-чужой»
- •Участки антигена, распознаваемые молекулами Участки антигена, распознаваемые молекулами главного комплекса гистосовместимости (МНС) и
- •Тримолекулярный комплекс
- •РаспознаваниеРаспознаваниеиипереработкапереработкаантигенантиген
- •Гуморальный иммунный ответ
- •КлональнаяКлональнаяселекцияселекцияВВ--клетокклеток
- •СтроениеСтроениеIgGIgG
- •Взаимодействие антигена и антитела
- •ОсновныеОсновныеформыформыпатологиипатологиииммуннойиммуннойсиси
- •ВВ--клеточныйклеточныйответответнанасобственныесобственные илииличужеродныечужеродныеантигеныантигены
- •ПерекрестноПерекрестно--реагирующиереагирующиеантигеныантигены индуцируютиндуцируютпоявлениепоявление аутоиммунныхаутоиммунныхТхТх--клетокклеток
- •ИндукцияИндукциясинтезасинтезааутоантителаутоантител перекрестноперекрестно--реагирующимиреагирующимиантигенамиантигенами
- •Лекция 2 Лекция 2
- •Insects and infections
- •Induction of antimicrobial activity by immune challenge
- •E. Metchnikow
- •ФагоцитарныеФагоцитарныеклеткиклетки..СистемаСистема мононуклеарныхмононуклеарныхфагоцитовфагоцитов
- •ХемотаксисХемотаксис
- •ХемотаксисХемотаксис
- •ХемокиныХемокины
- •ХемотаксисХемотаксис
- •ФагоцитозФагоцитоз
- •ФагоцитозФагоцитоз
- •Jules Hoffmann
- •drosophila
- •“Scavenger“ScavengerReceptors”Receptors”
- •СистемаСистемаTLRTLR
- •Toll
- •imd and Toll mutants are immunocompromised
- •How is infection sensed in insects?
- •The mutant semmelweis reveals
- •Sensing Gram-negative infections in Drosophila
- •Peptidoglycan structure Lys-type
- •Peptidoglycan structure
- •TCT/PGRP-LC complex
- •Drosophila recognition proteins for microbial structures
- •Fungi Yeast
- •Prizes won for discoveries in the field of TLRs
- •Mammalian TLR signalling pathways.
- •Innate immunity: sensing and signalling
- •Innate immunity and virus infection
- •Главные функции комплемента в воспалительном про
- •КлассическийКлассическийииальтернативныйальтернативный путипутиактивацииактивациисистемысистемы комплементакомплемента
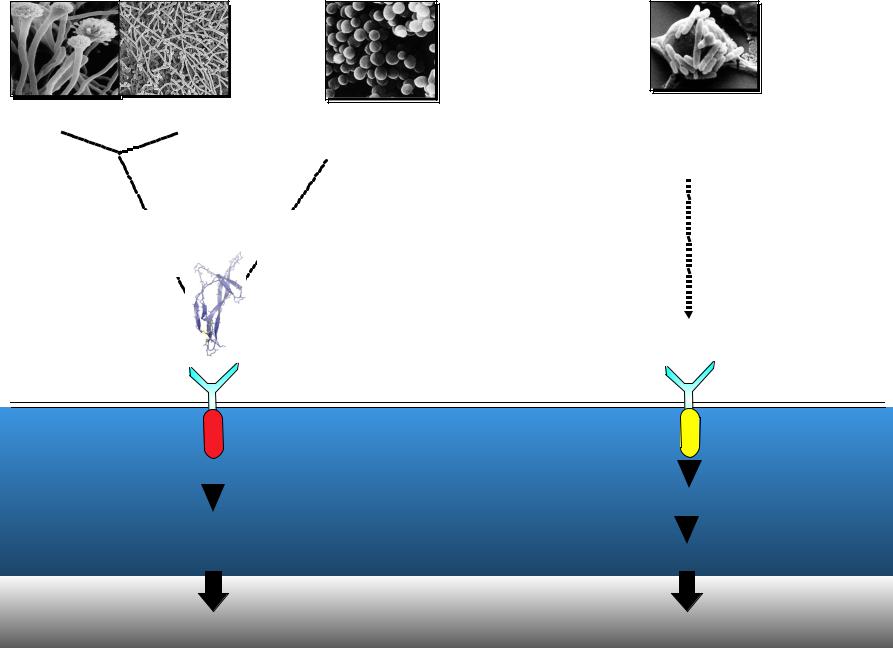
How is infection sensed in insects?
Fungi Yeast |
Gram positive |
Gram negative |
|
bacteria |
bacteria |
|
|
? |
? |
? |
|
Spz |
|
Toll |
|
Toll-2? |
|
|
IMD (RIP) |
Dif (NF- B ) |
Relish (NF- B ) |
|
Drosomycin |
Diptericin |
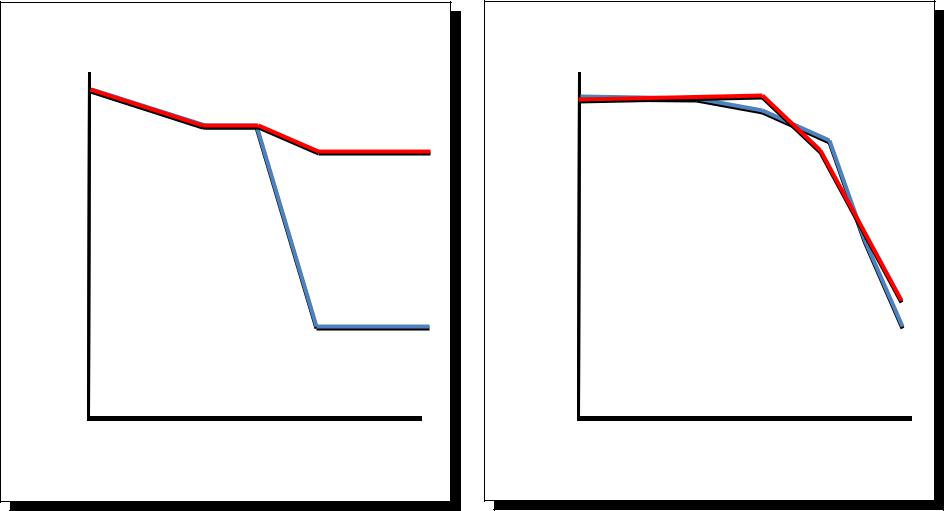
The mutant semmelweis reveals |
|
||||||||||
differences in the sensing of bacteria |
|||||||||||
|
|
|
and fungi |
|
|
|
|
|
|
||
Streptococcus faecalis |
|
Beauveria bassiana |
|
||||||||
100 |
(Gram +) |
|
Survival rate (%) |
100 |
|
(Fungus) |
|
|
|||
|
|
wt |
|
|
|
|
wt |
|
|||
|
|
|
|
|
|
|
|
|
|||
50 |
|
|
|
50 |
|
|
|
|
|
|
|
Survivalrate(%) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Seml |
|
|
|
|
|
|
Seml |
||
0 0 |
12 |
24 |
36 |
|
0 0 |
1 |
2 |
3 |
4 |
5 |
6 |
|
Time p.i. (h) |
|
|
|
|
Time p.i. (d) |
|
||||
Michel et al Nature (20

|
The PeptidoGlycan Recognition Protein |
|||||||||||||||||||||||||||||||||||||||||
|
|
|
|
|
|
|
(PGRP) family (drosophila) |
|||||||||||||||||||||||||||||||||||
|
SA |
|
|
|
Semmelweis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||||||||||||||
|
|
|
|
|
||||||||||||||||||||||||||||||||||||||
|
203 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||||||||||||||||||
|
SB1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
190 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
SB2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
182 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
SC1a |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
185 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
SC1b |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
185 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
SC2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
184 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||||||
|
SD |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
186 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||||
|
LA |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
280 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||||||
|
LB |
|
|
|
|
|
|
|
|
|
|
|
|
|
215 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
LC |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
520 |
|
||||||||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
LD |
|
|
|
|
|
|
|
|
|
|
|
|
|
505 |
|
||||||||||||||||||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
LE |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
345 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
LF |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
337 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Signal peptide |
PGRP domain with/without amidase |
Transmembrane domain |
activity |
|
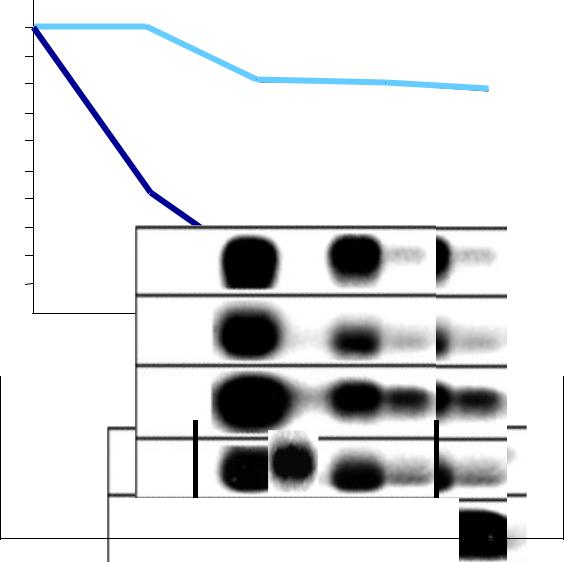
Sensing Gram-negative infections in Drosophila
Survival rate
infection by Enterobacter cloacae (Gram-)
100
PGRP-SA -/-
50
PGRP-LC -/-
12 |
|
24 |
36 |
|
48 |
|
|
Time (h) |
|
|
|
E. cloacae: - |
+ + + |
|
|
||
|
|
|
|
|
|
Diptericin |
|
|
|
|
|
|
|
|
|
||
|
|
|
- |
|
|
|
wt |
wt SA -/-LC -/ |
|
||
Gottar et al Nature (2

Peptidoglycan structure Lys-type
(most Gram+)
Glycan strand |
|
|
|
|
|
|
GlcNAc |
MurNAc |
GlcNAc |
MurNAc |
GlcNAc |
MurNAc |
|
|
L-Ala |
|
|
L-Ala |
|
L-Ala |
|
D-Glu |
|
|
D-Glu |
|
D-Glu |
Short peptide L-Lys |
(Gly) D-Ala |
|
|
L-Lys |
||
bridge |
D-Ala |
|
L-Lys |
|
|
D-Ala |
|
|
|
D-Glu |
|||
|
|
|
D-Glu |
|
|
|
|
|
|
L-Ala |
|
L-Ala |
|
Glycan strand |
|
MurNAc |
GlcNAc |
MurNAc |
|
|
|
GlcNAc |
|
||||

Peptidoglycan structure
Glycan strand
GlcNAc |
MurNAc |
GlcNAc |
MurNAc |
|
L-Ala |
|
|
|
|
|
L-Ala |
|||||
|
D- |
|
Glu |
|
|
|
|
|
D- |
|
Glu |
|
|
|
|
|
|
|
|
|
|||||
Short peptide DAP |
|
(Gly) |
|
D-Ala |
|
|
|
|||||
|
|
|
|
|||||||||
|
|
|
||||||||||
|
|
|
|
|
||||||||
bridge |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
D-Ala |
|
|
DAP |
|
|
|
||||||
|
|
|
|
|
|
|||||||
|
|
|
|
|
|
|
D- |
|
Glu |
|
|
|
|
|
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
L-Ala |
|
|
|
||
Glycan strand |
|
MurNAc |
GlcNAc |
|||||||||
|
GlcNAc |
|||||||||||
DAP-type
(Gram-)
TCT
GlcNAc MurNAc
L-Ala
D-Glu
DAP
D-Ala
D-Glu
L-Ala
MurNAc

TCT/PGRP-LC complex
DAP
From Chang et al (2006), Science 311: 17

Drosophila recognition proteins for microbial structures
Signal PGRP domain peptide
PGRP-SA |
|
|
|
|
|
|
|
|
(semmelweis) |
PGN (Ly |
|
|
|
|
|
|
|
|
|||
|
|
|
|
|
|
|
|
|||
|
|
|
180203 |
|
|
|||||
|
27 38 |
|
Michel et al (2001) Nat |
|||||||
|
|
|
|
|
Transmembrane |
|
||||
PGRP-LC |
|
PGRP domain |
|
|||||||
|
domain |
|
||||||||
295 317 |
353 |
PGN (DA |
498 520 |
Gottar et al (2002) Nat
GNBP-1
GNBP-3
GNBP homology |
|
|
|
-glucanase |
|
|
|
||||||||
|
|
|
|
|
domain |
|
|
|
domain |
|
|
PGN (Ly |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
19 |
117 |
194 |
|
447 |
492 |
||||||||
GNBP homology |
|
|
|
-glucanase |
Gobert et al (2003) Scie |
||||||||||
|
|
|
|
|
|
||||||||||
|
|
|
|
|
domain |
|
|
|
domain |
|
|
- ,3 gluc |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
26 |
121 |
242 |
466 |
492 |
||||||||||
Gottar et al (2006) C

Fungi Yeast |
Gram positive Gram negative |
Virus |
|||
|
|
bacteria |
bacteria |
|
|
-glucansl |
PGRP-SD |
Lys-PGN |
DAP-PGN |
|
|
|
PGRP-SA |
|
|
|
|
Microbial |
GNBP-3 |
|
|
|
|
|
|
|
|
|
|
protease |
|
GNBP- |
|
|
|
s |
|
|
|
|
? |
|
Spaetzle |
Toll |
PGRP-LC |
|
|
|
|
|
|||
MyD88 |
IMD (RIP) |
? |
|
||
|
Relish (NF- B ) |
|
Dif (NF- B ) |
|
|
Drosomycin |
Diptericin |
? |
Ferrandon et al (2007) Nat Rev Im

988 |
IL- R is cloned. |
989 |
Charles Janeway proposes the concept of pattern-recognition |
990 |
receptors. |
CD14 and LPS-binding protein are identified as components of the |
|
99 |
LPS receptor complex. |
Sequence similarity between Toll and IL-1R1 identified |
|
993– 996 |
Pathogen-specific immune signalling found to involve induction of |
|
antimicrobial peptides by members of the NF-κB family in Drosophila |
994 |
melanogaster. |
Plant protein N is shown to be involved in disease resistance and to |
|
996 |
have a TIR domain that is similar to Toll and IL-1R1. |
The Toll pathway is shown to regulate the antifungal response in D. |
|
997 |
melanogaster. |
The first human homologue of Toll receptor is cloned (hToll; later |
|
|
renamed TLR4). |
998 |
A role for MYD88 in IL-1 receptor signalling is identified. |
Four further human TLRs are identified. |
|
999 |
TLR4 is identified as the signalling receptor for LPS. |
LPS signalling is found to require MYD88. |
|
|
The requirement of MD2 for TLR4 responsiveness to LPS is identified. |
2000-2002 |
Ligands for TLR2- heterodimeric complexes are identified |
|
Viral antagonists of TLRs are identified. |
200 |
TLR9 is characterized as the receptor for CpG-DNA. |
The first TLR that recognizes viral components is identified (TLR3). |
|
|
Flagellin is identified as a ligand for TLR5. |
2002 |
MAL (also known as TIRAP) is discovered. |
TRIF is discovered. |
|
2002-2009 |
Endogenous ligands for TLRs are identified. |
2003 |
TRAM is discovered. |
2004 |
TLR7 and TLR8 are reported to recognize viral ssRNA. |
2006 |
The first function for mammalian SARM1 (a regulator of TRIF) is |
2007-2009 |
reported. |
Structures of several TLR–ligand complexes (including TLR4, TLR2– |
|
|
TLR1, TLR2–TLR6 and TLR3) are solved. |
