
4. Dna Polymerase
There are three enzymes, termed DNA polymerases, that are used to replicate DNA. Polymerase III copies the strand while I and II are responsible for proofreading and repairing the copied strand. These polymerase enzymes catalyze the addition of nucleotides to elongate the newly forming strand using the parent strand as a complementary template. A short chain of nucleotides, called a primer, is required by the polymerase to start. From this primer the rest of the strand can be formed. The enzymes can only add nucleotides in 5` to a 3` direction beginning with the 3` end of primer. The DNA polymerase catalyzes the nucleophilic attack of the 3`-OH from the growing strand on the α-phosphate of the nucleotide, releasing pyrophosphate. Thus, the polymerases can only add nucleotides.



5. Okazaki Fragments
Each strand of duplex DNA runs in an anti-parallel (opposite) direction to the other. One problem the replication machinery has to overcome is that the polymerase only travels in the 5` to 3` direction, yet must always move towards the replication fork. To solve this dilemma, the polymerase builds the lagging strand using many pieces, called Okazaki fragments, in a leapfrog fashion from one primer to the next. The leading strand simply follows in the direction of the replication fork.




6. Steps of dna replication
1. Unwinding the helix
The
first step in DNA replication is the formation of the replication
fork. The enzyme gyrase induces supercoiling to unwind the double
helix. Following the gyrase is helicase. Helicase breaks the hydrogen
bonds between the base pairs, allowing the parent strands to
separate. The helicase obtains the energy for this process by the
hydrolysis of adenosine triphosphate (ATP).
2. Stabilizing the Strands
The phosphodiesteric bonds of the single-stranded DNA are now vulnerable to attack through hydrolytic cleavage. Single-stranded DNA binding (SSB) proteins prevent these bonds being broken and keep the separated strands from reannealing the replication process.

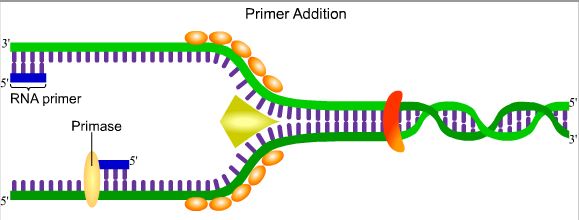
3. Primer addition
The
enzyme primase creates an RNA primer from which nucleotide
polymerization can begin. The lagging strand has the additional
complexity of the Okazaki fragments because of the 5` to 3`
polymerization requirement and also because overall polymerization
must move towards the replication fork.
4. Nucleotide Addition
After
the attachment of RNA primer, DNA polymerase III adds nucleotides to
the new strand in a 5` to 3` direction. Because the polymerization on
both strands must move towards the replication fork, the lagging
strand replication is more complex, requiring the use of Okazaki
fragments.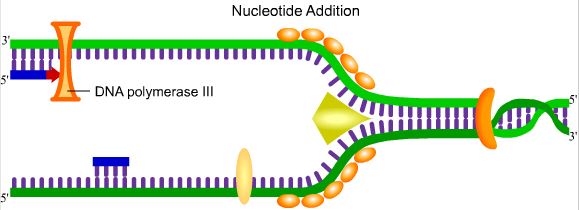

5. Primer Removal
The
RNA primers are then removed by DNA polymerase I, which also fills in
the resulting gaps with DNA. The polymerase is unable to connect the
3` nucleotide of the replacement nucleotides to the rest of the
strand, leaving a phosphodiesteric gap in the strand.

6. Filling the Gaps
Lastly,
the nicks between the Okazaki fragments on the lagging strand must be
joined. This is accomplished by the enzyme ligase. This reaction
requires the breakup of one ATP molecule into AMP and PPi.



